Abstract
Lung cancer is the leading cause of cancer mortality in Mexico and worldwide. In the past decade, there has been an increase in the number of lung cancer cases in young people, which suggests an important role for genetic background in the etiology of this disease. In this study, we genetically characterized 16 polymorphisms in 12 low penetrance genes (AhR, CYP1A1, CYP2E1, EPHX1, GSTM1, GSTT1, GSTPI, XRCC1, ERCC2, MGMT, CCND1 and TP53) in 382 healthy Mexican Mestizos as the first step in elucidating the genetic structure of this population and identifying high risk individuals. All of the genotypes analyzed were in Hardy-Weinberg equilibrium, but different degrees of linkage were observed for polymorphisms in the CYP1A1 and EPHX1 genes. The genetic variability of this population was distributed in six clusters that were defined based on their genetic characteristics. The use of a polygenic model to assess the additive effect of low penetrance risk alleles identified combinations of risk genotypes that could be useful in predicting a predisposition to lung cancer. Estimation of the level of genetic susceptibility showed that the individual calculated risk value (iCRV) ranged from 1 to 16, with a higher iCRV indicating a greater genetic susceptibility to lung cancer.
additive model; genetic polymorphism; lung cancer; Mexicans; molecular epidemiology
Polymorphisms associated with the risk of lung cancer in a healthy Mexican Mestizo population: Application of the additive model for cancer
Rebeca Pérez-MoralesI, * Send correspondence to: Julieta Rubio Departamento de Medicina Genómica y Toxicología Ambiental Instituto de Investigaciones Biomédicas Universidad Nacional Autónoma de México Apartado Postal 70228, 04510 Mexico City, México E-mail: juruli@unam.mx. ; Ignacio Méndez-RamírezII; Clementina Castro-HernándezI; Ollin C. Martínez-RamírezI; María Eugenia GonsebattI; Julieta RubioI
IDepartamento de Medicina Genómica y Toxicología Ambiental, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México, Mexico City, México
IIDepartamento de Probabilidad y Estadística, Instituto de Investigaciones en Matemáticas Aplicadas y en Sistemas, Universidad Nacional Autónoma de México, Mexico City, México
Send correspondence to Send correspondence to: Julieta Rubio Departamento de Medicina Genómica y Toxicología Ambiental Instituto de Investigaciones Biomédicas Universidad Nacional Autónoma de México Apartado Postal 70228, 04510 Mexico City, México E-mail: juruli@unam.mx.
ABSTRACT
Lung cancer is the leading cause of cancer mortality in Mexico and worldwide. In the past decade, there has been an increase in the number of lung cancer cases in young people, which suggests an important role for genetic background in the etiology of this disease. In this study, we genetically characterized 16 polymorphisms in 12 low penetrance genes (AhR, CYP1A1, CYP2E1, EPHX1, GSTM1, GSTT1, GSTPI, XRCC1, ERCC2, MGMT, CCND1 and TP53) in 382 healthy Mexican Mestizos as the first step in elucidating the genetic structure of this population and identifying high risk individuals. All of the genotypes analyzed were in Hardy-Weinberg equilibrium, but different degrees of linkage were observed for polymorphisms in the CYP1A1 and EPHX1 genes. The genetic variability of this population was distributed in six clusters that were defined based on their genetic characteristics. The use of a polygenic model to assess the additive effect of low penetrance risk alleles identified combinations of risk genotypes that could be useful in predicting a predisposition to lung cancer. Estimation of the level of genetic susceptibility showed that the individual calculated risk value (iCRV) ranged from 1 to 16, with a higher iCRV indicating a greater genetic susceptibility to lung cancer.
Key words: additive model, genetic polymorphism, lung cancer, Mexicans, molecular epidemiology.
Introduction
Lung cancer (LC) is the major cause of mortality from neoplasias worldwide. In Mexico, LC was responsible for 11.45% of deaths by malignant neoplasia from 1998-2002, and a prospective study indicated that mortality from LC will be even greater in the future (Ruíz-Godoy et al., 2007). Lung cancer is a serious public health problem (Proctor, 2001). The World Health Organization (WHO) estimates that in 2030 the number of deaths attributable to the consumption of tobacco will be 100 million, accompanied by an increased incidence of LC (Xie and Minna, 2008).
Lung cancer is highly related to tobacco consumption, although only 20% of smokers develop LC. On other hand, the number of cases among people under 50 years old has increased (Gemignani et al., 2007), highlighting the importance of genetic background in the etiology of LC. In this context, efforts have been made to identify polymorphisms associated with the risk of developing LC. Association studies have focused mainly on polymorphisms in genes coding for (1) enzymes involved in the bioactivation of carcinogens, such as cytochromes P450 CYP1A1 (Shah et al., 2008), CYP2E1 (Zhan et al., 2010) and CYP2D6 (Shaw et al., 1998), (2) enzymes involved in detoxification, e.g., glutathione transferases GSTM1, GSTT1 and GSTP1 (Sobti et al., 2008), N-acetyl transferase 2 (NAT2) and epoxide hydrolase (EPHX1) (Zhou et al., 2002), (3) proteins involved in DNA repair, such as XRCC1, XRCC3, ERCC2, ERCC4, MGMT and OGG1 (Zienolddiny et al., 2006; Hung et al., 2007) and (4) proteins implicated in the cell cycle and apoptosis, such as CCND1, CHEK2, MDM2, TP53 and EGF (Hosgood III et al., 2008).
In some populations, a significant association between gene polymorphisms and the risk of LC risk has been established, while in other populations no associations have been found, probably because of the low frequency of polymorphisms. For example, the CYP1A1 rs1048943 (Ile462Val) polymorphism is associated with a risk of LC in Asian Korean, Chinese and Japanese populations (Lee et al., 2008) and in Chilean (Quinoñes et al., 2001), Mexican (Gallegos-Arreola et al., 2008) and Afro-American (Taioli et al., 1998) populations, whereas this association has not been confirmed in American and European Caucasians, possibly because the frequency of the CYP1A1 rs1048943 polymorphism in these populations is < 2% (Hung et al., 2003). In contrast, the GSTM1 deletion polymorphism, which has a frequency of 0.35-0.58 in Asians, European and American Caucasians and Africans (Mohr et al., 2003), is significantly associated with LC in Caucasians (Hung et al., 2003) and Asians (Lee et al., 2008). ERCC2 rs1799793 (Asp399Asn), ERCC2 rs13181 (Lys751Asn) and MGMT rs12917 (Leu84Phe) polymorphisms show no consistent association with LC in different populations (Kiyohara and Yoshimasu, 2007). However, XRCC1 rs3213245 (-77 T > C) was associated with a risk of LC in three case-control studies and ERCC2 rs13181 was associated with a risk of LC in a meta-analysis study of 18 case-control reports (Vineis et al., 2009).
Since these polymorphisms modify the functionality of the encoded proteins it has been suggested that polymorphic variants may alter the metabolic and detoxification pathways of carcinogenic compounds, thereby predisposing the individual bearing these polymorphisms to develop cancer (Nebert and Dalton, 2006). Indeed, polymorphisms in specific genes can modulate the formation of DNA-carcinogen adducts (Ketelslegers et al., 2006) which favors the generation of mutations leading to LC, particularly in smokers (Lodovici et al., 2004).
Recently, a polygenic cancer model was proposed that considers the genetic susceptibility to cancer as a global mechanism, with the susceptibility being defined by low risk alleles in multiple candidate genes (Pharoah et al., 2002; Dong et al., 2008). Susceptibility to LC may be caused by low penetrance genes (low risk) with high frequencies in the general population (Kiyohara et al., 2007). In this context, susceptibility to LC is determined by a combination of multiple low risk alleles in an individual, with each allele contributing only slightly to the overall cancer risk, as proposed by Fletcher and Houlston (2010) in their polygenic additive model. This model, which allows the identification of high risk individuals, may be useful in preventing LC in the early stages, thereby significantly reducing LC-related mortality and the costs associated with the diagnosis and treatment of this disease.
The first step in any study of molecular epidemiology in which ethnicity plays an important role is the characterization of the general healthy population since this will provide the benchmark for further analysis. In this study, we investigated 16 polymorphisms in 12 low penetrance genes in a healthy Mexican Mestizo population. These genes code for proteins involved in the metabolic pathways of some environmental and tobacco smoke carcinogens, with their polymorphisms reportedly producing functional alterations that are associated with the risk of developing LC. The association between these polymorphisms and the risk of cancer was assessed using the polygenic additive model for cancer.
Materials and Methods
Subjects
The research protocol was approved by the Committee of Bioethics of the Instituto de Investigaciones Biomédicas of the Universidad Nacional Autónoma de México, and the Hospital "20 de Noviembre" ISSSTE gave permission to use the buffy coat of blood bank samples as a source of DNA. The study included 382 unrelated, healthy Mexican Mestizo individuals whose parents and grandparents were born in Mexico. After providing informed consent, the subjects answered a questionnaire that included information on their age, gender, smoking status and lifestyle.
Polymorphism analysis
Genomic DNA was obtained from blood samples by phenol-chloroform extraction and ethanol precipitation (Sambrook et al., 1987). The presence of the AhR rs2066853 (Arg554Lys) polymorphism was determined by using the TaqMan probe C_11170747_20 from Applied Biosystems, according to the manufacturer's recommendations. Restriction fragment length polymorphism (RFLP) analysis was used to assess the following polymorphisms: CYP1A1 rs4646903 (6235T > C) and CYP1A1 rs1800031 (5639T > C) (Kawajiri, 1999), CYP1A1 rs1048943 (Ile462Val) and CYP1A1 rs1799814 (Thr461Asn) (Cascorbi et al., 1996), CYP2E1 rs2031920 (-1053C > T) (Morita et al., 1997), EPHX1 rs1051740 (Tyr113His) and EPHX1 rs2234922 (His139Arg) (Smith and Harrison, 1997), GSTPI rs947894 (Ile105Val) (Harries et al., 1997), XRCC1 rs25487 (Arg399Gln) (Abdel-Rahman and El-Zein, 2000), ERCC2 rs13181 (Lys751Gln) (Lunn et al., 2000), MGMT rs12917 (Leu84Phe) (Courtney et al., 2005), CCND1 rs603965 (870G > A) (Wang et al., 2002), and TP53 rs1042522 (Arg72Pro) (Irarrázabal et al., 2003). Polymorphisms of GSTT1 null and GSTM1 null were determined by multiplex PCR using a previously reported protocol (Abdel-Rahman et al., 1996).
Statistical analyses
The statistical package GenePop version 4.0.10 (http://genepop.curtin.edu.au) was used to assess whether the genotypes of each gene were in Hardy-Weinberg equilibrium and to determine the degree of linkage between the EPHX1 and CYP1A1 gene polymorphisms. Conglomerate and hierarchical clustering analyses were used to determine the genetic variability of the sample. The estimated cancer risk and the genotypic and allelic frequencies were determined using the statistical package JMP version 8.
Results
Of the 382 Mexican Mestizo individuals studied, 29% were women and 71% were men. The age range was between 18 and 80 years (mean age: 39.2 ± 12.1 for men and 41.5 ± 13.1 for women) and 48% of the population were smokers.
The following polymorphisms were studied in candidate genes: AhR rs2066853, CYP1A1 rs4646903, CYP1A1 rs1048943, CYP1A1 rs1800031, CYP1A1 rs1799814, CYP2E1 rs2031920, EPHX1 rs1051740, EPHX1 rs2234922, GSTM1 null, GSTT1 null, GSTPI rs947894, XRCC1 rs25487, ERCC2 rs13181, MGMT rs12917, CCND1 rs603965 and TP53 rs1042522. The genotypic and allelic frequencies of these polymorphisms are shown in Table 1. All genotypes were in Hardy-Weinberg equilibrium. GSTM1 and GSTT1 were not analyzed for Hardy-Weinberg equilibrium because the methodology did not allow discrimination between heterozygous and homozygous positive genotypes.
Polymorphisms in the same gene may have synergistic or antagonistic effects, as in the case of CYP1A1 and epoxide hydrolase 1. This is functionally significant because CYP1A1 acts in association with EPHX1 to convert polyaromatic hydrocarbons to highly toxic, mutagenic and carcinogenic epoxides.
Linkage analysis of four CYP1A1 and two EPHX1 polymorphisms showed that the CYP1A1 rs1799814 and CYP1A1 rs1800031 genotypes were linked in all cases, with a probability of 1 (p < 0.001), whereas CYP1A1 rs4646903 and CYP1A1 rs1048943 polymorphisms were not linked, with a probability of 0 (p < 0.001). EPHX1 rs1051740 and EPHX1 rs2234922 were linked, with a probability of 0.03254 (p = 0.0025). Combinations of CYP1A1 polymorphisms are shown in Table 2.
Conglomerate analysis was used to determine the genetic variability and the possible grouping of the individuals analyzed. We identified six groups that clustered according to their genetic characteristics, although there was considerable heterogeneity (Figure 1). Additionally, hierarchical cluster analysis of the genotypes showed that CYP1A1 rs1800031 and CYP1A1 rs1799814 polymorphisms clustered together, whereas CYP1A1 rs4646903 and CYP1A1 rs1048943 were close to each other but clustered separately (Figure 2). The EPHX1 rs1051740 and EPHX1 rs2234922 polymorphisms were widely separated, as also indicated by linkage analysis.
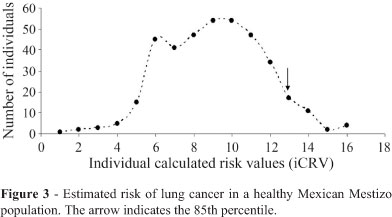
To determine the theoretical levels of susceptibility to LC, a risk matrix was generated using a log-additive model in which a value of 0 was assigned to homozygous genotypes that produced no risk, 1 to heterozygous genotypes (medium risk) and 2 to homozygous genotypes that produced changes in the activity of the protein, considered to be high risk. The individual calculated risk values (iCRVs) were determined by adding the values of the log-additive model to each locus. The iCRVs ranged from 1 to 16, although there was a marked decrease in individuals with iCRV < 5 and > 12 (Figure 3).
Discussion
Cancer is a polygenic disease, the risk of which may be related to the presence of low-penetrance genes that have additive effects. In this study, we examined the frequency of some polymorphisms possibly related to the risk of developing lung cancer in a sample of healthy Mexican Mestizos. These polymorphisms may be useful biomarkers of genetic susceptibility to lung cancer in specific populations.
We have previously reported on the high frequency of the CYP1A1 rs1048943 polymorphism in Mexican Mestizos (Pérez-Morales et al., 2008), a polymorphism that is highly represented in Amerindians (Kvitko et al., 2000). In some populations, the CYP1A1 rs1048943 polymorphism has been associated with a risk of LC (Quinoñes et al., 2001; Lee et al., 2008; Shah et al., 2008), although not all studies have found such an association, especially when the frequency of this polymorphism is very low, as in European and American Caucasian populations (Hung et al., 2003). However, in some populations the CYP1A1 rs1048943 polymorphism has a significant influence on the risk of developing LC because of the additive effect of other polymorphisms.
Although the CYP1A1 rs1799814 and CYP1A1 rs1800031 genotypes were found to be linked, the linkage analysis may have been affected by the high frequencies of the wild type allele at these loci in our population. On the other hand, in contrast to a previous report (Hayashi et al., 1991), the CYP1A1 rs4646903 and CYP1A1 rs1048943 polymorphisms were not linked. This observation is functionally significant because the CYP1A1 rs1048943 polymorphism increases the activity of cytochrome CYP1A1, resulting in a more efficient generation of reactive metabolites, whereas the CYP1A1 rs4646903 polymorphism increases the levels of CYP1A1 mRNA. Hence, the combination of these alleles could increase the risk of LC (Yoon et al., 2008). Some studies have associated CYP1A1 rs1799814 polymorphism with a risk of LC (Gallegos-Arreola et al., 2008), whereas CYP1A1 rs1800031 is reportedly specific for African populations but is not associated with a risk of LC (Taioli et al., 1998).
The proportion of the population carrying both the EPHX1 rs1051740 and EPHX1 rs2234922 alleles was very small. In these individuals, the allele EPHX1 rs1051740 Tyr113 apparently offered no protection against LC, in contrast to previous observations (Voho et al., 2006). On the other hand, the EPHX1 rs2234922 139Arg variant increases the activity of the encoded enzyme (Hassett et al., 1994), which could suppress the low activity of the EPHX1 113His variant (Salam et al., 2007). For example, an individual with the CYP1A1 rs4646903, CYP1A1 rs1048943, EPHX1 rs1051740 and EPHX1 rs2234922 polymorphisms will produce reactive metabolites more efficiently and consequently have a higher risk of developing LC.
A conglomerate analysis of the genetic variability of the population studied here revealed six groups that clustered according to their genetic characteristics (Figure 1). In clusters 2 and 4, the polymorphism CYP1A1 rs1800031 was quite separate from the rest of the genotypes, although it should be noted that these two groups consisted of only two subjects each such that the probability of finding individuals belonging to these clusters in a given population is very low. In clusters 2, 3, 4 and 5, AhR rs2066853, EPHX1 rs2234922, GSTT1 null, ERCC2 rs13181, and CYP1A1 rs1799814 were over-represented with respect to the other polymorphisms analyzed, but the number of individuals was smaller than in clusters 1 and 6; there was no predominant genotype in the latter two clusters because of the combination of polymorphic alleles at each locus in each individual. Despite this clustering, our findings indicate that the population studied was highly heterogenous, as is characteristic of Mexican Mestizos.
Hierarchical cluster analysis showed that the genes were grouped according to their frequency. This analysis also revealed that CYP1A1 rs1800031 and CYP1A1 rs1799814 occurred together, in agreement with the linkage analysis, whereas CYP1A1 rs4646903 and CYP1A1 rs1048943 were found together in some individuals but were not linked (as indicated by the separation of their branches). Overall, this analysis showed that a portion of the population carried both risk alleles, although they were not linked. However, we have not determined whether these polymorphisms are in a cis or trans position, which could affect the linkage results.
Our analysis revealed individuals with a high susceptibility to LC based on the presence of risk genotypes, although different combinations of risk genotypes may confer varying degrees of susceptibility when combined with other components, such as environmental factors.
If the risk of developing LC is attributed to the interaction of several low-penetrance genes that exert an additive effect then we should be able to detect individuals with a high number of risk alleles and a greater genetic susceptibility to LC. We estimated the levels of susceptibility and found that the iCRV ranged from 1 to 16; there was a marked decrease in the susceptibility to LC among individuals with an iCRV > 12. Application of the polygenic model (Fletcher and Houlston, 2010) (Figure 3) yielded a normal distribution of risk alleles in which low and high risk individuals occurred at the extremes of the distribution. Theoretically, high-risk individuals should be more susceptible to LC, but one cannot exclude the important role of genotype x environmental interactions to which individuals are exposed.
Based on the polygenic model of cancer, which takes into consideration the additive effect of multiple risk alleles, the high risk genotypes identified in this study included genes involved in phase I and II metabolism, DNA repair, oxidative stress and cell cycle regulation. All of these gene groups need to be considered when analyzing the efficiency of reactive metabolite generation, DNA-adduct repair, damage persistence and the cellular response to cycle arrest or apoptosis. However, as shown here, only a small minority of individuals actually possess a large number of risk alleles. Although studies of other populations have associated specific polymorphisms with a risk of LC, these relationships have not always been confirmed, possibly because of the pleiotropic or epistatic effects of one genotype on another within the same population.
Since LC is a multifactorial, polygenic disease, it is incorrect to attribute the susceptibility to LC to a single gene or group of related genes. In this context, determining the genetic background of a healthy population, such as done here for a Mexican Mestizo population, can provide a sound basis for subsequent studies on the association between these risk genotypes and LC. In such cases, the iCRV should be higher in patients than in the healthy controls.
Acknowledgments
This study was supported by a grant (PAPIIT-IN219807) from the Universidad Nacional Autónoma de México to J.R. and a post-graduate scholarship (no. 204279) from the Consejo Nacional de Ciencia y Tecnología to R.P. We thank Dr. Sergio García Méndez, head of the Blood Bank, Hospital "20 de Noviembre" ISSSTE in Mexico City, for providing the samples, and Dr. Pavel Petrosyan for critical reading of the manuscript.
Internet Resources
Received: February 3, 2011
Accepted: July 18, 2011.
Associate Editor: Emmanuel Dias Neto
License information: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
- Abdel-Rahman S and El-Zein R (2000) The 399Gln polymorphism in the DNA repair gene XRCC1 modulates the genotoxic response induced in human lymphocytes by the tobacco-specific nitrosamine NNK. Cancer Lett 159:63-71.
- Abdel-Rahman S, El-Zein R, Anwar W and Au W (1996) A multiplex PCR procedure for polymorphic analysis of GSTM1 and GSTT1 genes in population studies. Cancer Lett 107:229-233.
- Cascorbi I, Brockmöller J and Roots I (1996) A C4887A polymorphism in exon 7 of human CYP1A1: Population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res 56:4965-4969.
- Courtney E, Wickliffe J, Wolfe K, Kinslow C, Lopez M and Abdel-Rahman S (2005) The L84F and the I143V polymorphism in the O6-methylguanine-DNA-methyltransferase (MGMT) gene increase human sensitivity to the genotoxic effects of the tobacco-specific nitrosamine carcinogen NNK. Pharmacogenet Genomics 15:571-578.
- Dong L, Potter J, White E, Ulrich C, Cardon L and Peters U (2008) Genetic susceptibility to cancer: The role of polymorphism in candidate genes. JAMA 299:2423-2434.
- Fletcher O and Houlston R (2010) Architecture of inherited susceptibility to common cancer. Nat Rev Cancer 10:353-361.
- Gallegos-Arreola M, Figuera-Villanueva L, Troyo-Sanroman R, Morgan-Villela G, Puebla-Pérez AM, Flores Márquez MR and Zúñiga-González GM (2008) CYP1A1*2B and *4 polymorphism are associated with lung cancer susceptibility in Mexican patients. Int J Biol Markers 23:1-7.
- Gemignani F, Landi S, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, et al. (2007) Development of lung cancer before the age of 50: The role of xenobiotic metabolizing genes. Carcinogenesis 28:1287-1293.
- Harries L, Stubbins M, Forman D, Howard G and Wolf R (1997) Identification of genetic polymorphism at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis 18:641-644.
- Hassett C, Aicher L, Sidhu J and Omiecinski C (1994) Human microsomal epoxide hydrolase: Genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet 3:421-428.
- Hayashi S, Watanabe J, Nakachi K and Kawajiri K (1991) Genetic linkage of lung cancer-associated MspI polymorphism with amino acid replacement in the heme binding region of the human cytochrome P450IA1. J Biochem 110:407-411.
- Hosgood III H, Menashe I, Shen M, Yeager M, Yuenger J, Rajaraman P, He X, Chatterjee N, Caporasso N, Zhu Y, et al. (2008) Pathway-based evaluation of 380 candidate genes and lung cancer susceptibility suggests the importance of the cell cycle pathway. Carcinogenesis 29:1938-1943.
- Hung R, Boffeta P, Brockmöller J, Haugen A, Butkiewicz D, Cascorbi I, Clapper M, Garte S, Hirvonen A, Anttila S, et al. (2003) CYP1A1 and GSTM1 genetic polymorphism and lung cancer risk in Caucasian non-smokers: A pooled analysis. Carcinogenesis 24:875-882.
- Hung R, Baragatti M, Thomas D, McKay J, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, et al. (2007) Inherited predisposition of lung cancer: A hierarchical modeling approach to DNA repair and cell cycle control pathways. Cancer Epidemiol Biomarkers Prev 16:2736-2744.
- Irarrázabal C, Rojas C, Aracena R, Márquez C and Gil L (2003) Chilean pilot study on the risk of lung cancer associated with codon 72 polymorphism in the gene of protein p53. Toxicol Lett 144:69-76.
- Kawajiri K (1999) Metabolic polymorphism and susceptibility to cancer. Chapter 15 CYP1A1 IARC Sci Publ 148:159-172.
- Ketelslegers H, Gottschalk R, Godschalk R, Knaapen AD, Schooten F, Vlietnick F, Kleinjans J and Van Delft J (2006) Interindividual variations in DNA adduct levels assessed by analysis of multiple genetic polymorphism in smokers. Cancer Epidemiol Biomarkers Prev 15:624-629.
- Kiyohara C and Yoshimasu K (2007) Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: A meta-analysis. Int J Med Sci 4:59-71.
- Kiyohara C, Yoshimasu K, Takayama K and Nakanishi Y (2007) Lung cancer susceptibility: Are we on our way to identifying a high-risk group? Future Oncol 3:617-627.
- Kvitko K, Nunes J, Weimer T, Salzano F and Hutz M (2000) Cytochrome P4501A1 polymorphism in South American Indians. Hum Biol 72:1039-1043.
- Lee K, Kang D, Clapper M, Ingelman-Sundberg M, Ono-Kihara M, Kiyohara C, Min S, Lan Q, Le Marchand L, Lin P, et al. (2008) CYP1A1, GSTM1, and GSTT1 polymorphisms, smoking, and lung cancer risk in a pooled analysis among Asian populations. Cancer Epidemiol Biomarkers Prev 17:1120-1126.
- Lodovici M, Luceri C, Guglielmi F, Bacci C, Akpan V, Fonnesu M, Boddi V and Dolara P (2004) Benzo(a)pyrene diolepoxide (BPDE)-DNA adduct levels in leukocytes of smokers in relation to polymorphism of CYP1A1, GSTM1, GSTP1, GSTT1 and mEH Cancer Epidemiol Biomarkers Prev 13:1342-1348.
- Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, Sanford KK and Bell DA (2000) XPD polymorphism: Effects on DNA repair proficiency. Carcinogenesis 21:551-555.
- Mohr L, Rodgers K and Silvestri G (2003) Glutathione S-transferase M1 polymorphism and the risk of lung cancer. Anticancer Res 23:2111-2124.
- Morita S, Yano M, Shiozaki H, Tsujinaka T, Ebisui C, Morimoto T, Kishibuti M, Fujita J, Ogawa A and Taniguchi M (1997) CYP1A1, CYP2E1 and GSTM1 polymorphism are not associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer 71:192-195.
- Nebert D and Dalton T (2006) The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer 6:947-960.
- Pérez-Morales R, Castro-Hernández C, Gonsebatt M and Rubio J (2008) Polymorphism of CYP1A1*2C, GSTM1*0, and GSTT1*0 in a Mexican Mestizo population: A similitude analysis. Hum Biol 80:457-465.
- Pharoah P, Antoniou A, Bobrow M, Zimmern R, Easton D and Ponder B (2002) Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet 31:33-36.
- Proctor R (2001) Tobacco and the global lung cancer epidemic. Nat Rev Cancer 1:82-86.
- Quinoñes L, Lucas D, Godoy J, Cáceres D, Berthou F, Varela N, Lee K, Acevedo C, Martínez L, Aguilera A, et al. (2001) CYP1A1, CYP2E1 and GSTM1 genetic polimorphisms. The effect of single and combined genotypes on lung cancer susceptibility in Chilean people. Cancer Lett 174:35-44.
- Ruíz-Godoy L, Rizo P, Sánchez F, Osornio-Vargas A, García-Cuellar C and Meneses A. Mortality due to lung in Mexico. Lung Cancer 58:184-190.
- Salam M, Lin P, Avol E, Gauderman J and Gilliland F (2007) Microsomal epoxide hydrolase, glutathione S-transferase P1, traffic and childhood asthma. Thorax 62:1050-1057.
- Sambrook J, Fritsch EF and Maniatis T (1987) Molecular Cloning. A Laboratory Manual. 2nd edition. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- Shah P, Singh A, Singh M, Mathur N, Pant M, Mishra B and Parmar D (2008) Interaction of cytochrome P4501A1 genotypes with other risk factors and susceptibility to lung cancer. Mutat Res 639:1-10.
- Shaw G, Falk R, Frame J, Weiffenbach B, Nesbitt J, Pass H, Caporaso N, Moir D and Tucker M (1998) Genetic polymorphism of CYPD6 and lung cancer risk. Cancer Epidemiol Biomarker Prev 7:215-219.
- Smith C and Harrison D (1997) Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema. Lancet 350:630-633.
- Sobti R, Kaur P, Kaur S, Janmeja A, Jindal S, Kishan J and Raimondi S (2008) Combined effect of GSTM1, GSTT1, and GSTP1 polymorphism on histological subtypes of lung cancer. Biomarkers 13:282-295.
- Taioli E, Ford J, Trachman J, Li Y, Demopoulos R and Garte S (1998) Lung cancer risk and CYP1A1 genotype in African Americans. Carcinogenesis 19:813-817.
- Vineis P, Manuguerra M, Kavvoura F, Guarrera S, Allione A, Rosa F, Di Gregorio A, Polidoro S, Saletta F, Ioannidis J, et al. (2009) A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst 101:24-36.
- Voho A, Metsola K, Anttila S, Impivaara O, Järvisalo J, Vainio H, Husgafvel-Pursiainen K and Hirvonen A (2006) EPHX1 gene polymorphism and individual susceptibility to lung cancer. Cancer Lett 237:102-108.
- Wang L, Habuchi T, Takahashi T, Mitsumori K, Kamamoto T, Kakei Y, Kakinuma H, Sato K, Nakamura A, Ogawa O, et al. (2002) Cyclin D1 gene polymorphism is associated with an increased risk of urinary bladder cancer. Carcinogenesis 23:257-264.
- Xie Y and Minna J (2008) Predicting the future for people with lung cancer. Nat Med 14:812-813.
- Yoon K, Kim J, Gil H, Hwang H, Hwangbo B and Lee J (2008) CYP1B1, CYP1A1, MPO, and GSTP1 polymorphism and lung cancer in never-smoking Korean women. Lung Cancer 60:40-46.
- Zhan P, Wang J, Zhang Y, Qiu L, Zhao S, Qian Q, Wei S, Yu L and Song Y (2010) CYP2E1 Rsa I/Pst I polymorphism is associated with lung cancer risk among Asians. Lung Cancer 69:19-25.
- Zhou W, Liu G, Thurston S, Xu L, Miller D, Wain J, Lynch T, Su L and Christiani DC (2002) Genetic polymorphism in N-acetyltransferase-2 and microsomal epoxide hydrolase, cumulative cigarette smoking, and lung cancer. Cancer Epidemiol Biomarker Prev 11:15-21.
- Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F and Haugen A (2006) Polymorphism of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 27:560-567.
- GenePop statistical package, http://genepop.curtin.edu.au (October 5, 2010).
» link
Publication Dates
-
Publication in this collection
03 Nov 2011 -
Date of issue
2011
History
-
Received
03 Feb 2011 -
Accepted
18 July 2011