Abstract
The cytogenetic characteristics of Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum and their F1, F2 and backcross hybrids were assessed by using chromosome banding techniques. The diploid number of 56 chromosomes was constant in all species and lineages, with a karyotypic formula containing 20 metacentric, 12 submetacentric, 12 subtelocentric and 12 acrocentric chromosomes. Nucleolar organizer regions (NORs) were identified in two subtelocentric chromosomes in the parents and hybrids, with partial nucleolar dominance in F1 and F2 specimens. Heterochromatic blocks were detected in the terminal and centromeric regions of some chromosomes in all individuals. For parental and hybrid lineages, 18S ribosomal clusters corresponding to NORs and 5S ribosomal genes were identified in distinct pairs of chromosomes. The striking conservation in the chromosomal macrostructure of the parental species may account for the fertility of their F1 hybrids. Similarly, the lack of marked alterations in the chromosomal structure of the F1 hybrids could account for the maintenance of these features in post-F1 lineages.
backcrossing; cytogenetics; interspecific hybrids; post-F1 hybrids; Pseudoplatystoma
Cytogenetic characterization of F1, F2 and backcross hybrids of the Neotropical catfish species Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum (Pimelodidae, Siluriformes)
Fernanda Dotti do PradoI; Tatiana Leite NunesII; José Augusto SenhoriniIII; Jehud BortolozziI; Fausto ForestiII; Fábio Porto-ForestiI
IDepartamento de Ciências Biológicas, Faculdade de Ciências, Universidade Estadual Paulista "Júlio de Mesquita Filho", Bauru, SP, Brazil
IIDepartamento de Morfologia, Instituto de Biociências, Universidade Estadual Paulista "Júlio de Mesquita Filho", Botucatu, SP, Brazil
IIICentro Nacional de Pesquisa e Conservação de Peixes Continentais, Pirassununga, SP, Brazil
Send correspondence to Send correspondence to: Fabio Porto Foresti Departamento de Ciências Biológicas Faculdade de Ciências Universidade Estadual Paulista "Júlio de Mesquita Filho" Av. Eng. Luiz Edmundo Carrijo Coube 14-01 17033-360 Bauru, SP, Brazil E-mail: fpforesti@fc.unesp.br
ABSTRACT
The cytogenetic characteristics of Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum and their F1, F2 and backcross hybrids were assessed by using chromosome banding techniques. The diploid number of 56 chromosomes was constant in all species and lineages, with a karyotypic formula containing 20 metacentric, 12 submetacentric, 12 subtelocentric and 12 acrocentric chromosomes. Nucleolar organizer regions (NORs) were identified in two subtelocentric chromosomes in the parents and hybrids, with partial nucleolar dominance in F1 and F2 specimens. Heterochromatic blocks were detected in the terminal and centromeric regions of some chromosomes in all individuals. For parental and hybrid lineages, 18S ribosomal clusters corresponding to NORs and 5S ribosomal genes were identified in distinct pairs of chromosomes. The striking conservation in the chromosomal macrostructure of the parental species may account for the fertility of their F1 hybrids. Similarly, the lack of marked alterations in the chromosomal structure of the F1 hybrids could account for the maintenance of these features in post-F1 lineages.
Key words: backcrossing, cytogenetics, interspecific hybrids, post-F1 hybrids, Pseudoplatystoma.
Introduction
The genetic identification of hybrids is an important aspect of fish culture that helps to avoid problems associated with artificial hybridization (Toledo-Filho et al., 1998; Porto-Foresti and Foresti, 2004). Among the currently available genetic methods, cytogenetics has contributed extensively to ichthyological studies and is an important tool for chromosomal, evolutionary and phylogenetic analyses and species identification (Foresti, 2008; Oliveira et al., 2009). In hybridization studies, this technique is effective in assessing the levels of ploidy and in identifying gynogenetic and androgenetic lineages, the morphological and chromosomal structure of parental and hybrid species and lineages, structural alterations in the chromosomes of hybrids, and cytogenetic markers that allow discrimination between pure species and interspecific hybrid lineages (Toledo-Filho et al., 1994; Almeida-Toledo et al., 1995; Porto-Foresti et al., 2008).
Fishes of the genus Pseudoplatystoma, such as P. corruscans (pintado) and P. reticulatum (cachara), have been used in large scale interspecific hybridization programs in Brazil (Godinho, 2007; Campos, 2010). These species belong to the family Pimelodidae, an important group of Neotropical Siluriformes (Lundberg and Littmann, 2003; Ferraris Jr, 2007) with an ample Neotropical distribution, and occur in the Amazon River basin (P. reticulatum), San Francisco River basin (P. corruscans) and Prata River basin (P. corruscans and P. reticulatum) (Buitrago-Suarez and Burr, 2007).
Both of the reciprocal interspecific hybrids of these species, denominated "cachapinta" (produced by crossing cachara females with pintado males) and "pintachara" (produced by crossing pintado females and cachara males) are viable, commercially advantageous, and have good market acceptance (Godinho, 2007; Porto-Foresti et al., 2008). These hybrids are fertile and are able to cross and produce post-F1 hybrids (F2 hybrids and backcrosses) in lineage cultivation, mainly involving the hybrid "cachapinta".
Despite the fact that these hybrids are economically important and have been extensively raised in fish-farming environments, there are no chromosomal data for these lineages. The aim of this study was therefore to use chromosomal banding techniques to cytogenetically characterize F1 interspecific hybrids, F2 hybrids and individuals resulting from backcrosses involving P. corruscans and P. reticulatum, in order to assess potential differences that could be used to characterize the parental species and identify hybridization products.
Material and Methods
21 specimens of P. corruscans and 15 of P. reticulatum were used as the parental lineages. We also studied 15 specimens of the hybrid "pintachara" and 16 of the hybrid "cachapinta" as representing the F1 interspecific hybrid lineage, in addition to five F2 specimens from the "cachapinta" intercross and 10 backcross individuals from backcrosses involving a "cachapinta" female and a pintado male; the latter two groups belonging to a post-F1 hybrid lineage.
All of the specimens were obtained from the hatchery stocks of the Centro Nacional de Pesquisa e Conservação de Peixes Continentais do Instituto Chico Mendes da Conservação de Biodiversidade (CEPTA/ICMBio, Pirassununga, SP, Brazil), which is also where the hybrids were artificially produced. The low number of F2 hybrids analyzed reflected the high mortality during the first weeks of stockpiling.
The samples of P. corruscans and P. reticulatum were collected throughout the Paraguay River basin; additional samples of P. corruscans were also collected from the Mogi-Guaçu River (Upper Paraná River basin). The fish were identified and stocked in the collection of the Laboratório de Genética de Peixes, UNESP, Bauru, SP, Brazil.
Mitotic chromosomes were obtained by initially injecting the fish with a biological fermentation solution (Lozano et al., 1988; Oliveira et al., 1988) to stimulate mitosis, after which a kidney cell suspension was used to provide chromosomal preparations, essentially as described by Foresti et al. (1981). Constitutive heterochromatin was detected by the C-banding technique described by Sumner (1972) and nucleolar organizer regions (NORs) were identified by silver nitrate staining (Ag-NOR), according to Howell and Black (1980).
The 18S and 5S ribosomal genes were identified simultaneously by fluorescent in situ hybridization (FISH) using 18S and 5S rDNA (ribosomal DNA) probes. The probes were obtained by polymerase chain reaction (PCR) amplification using the primers NS1 (5'-GTAGTCATATGCTTGTCTC-3') and NS8 (5'-TCCGCAGGTTCACCTACGGA-3') for the 18S gene (White et al., 1990) and primers A (5'-TACGCCCGATC TC GTCCGATC-3') and B (5'-CAGGCTGGTATGGCC GTAAGC-3') for the 5S gene (Pendás et al., 1994). The probes were labelled with biotin-dATP (18S) and digoxigenin-dUTP (5S) by nick translation, according to the manufacturer's instructions (Bionick labelling system, Gibco BRL). After labeling, the hybridizations were done using the technique described by Pinkel et al. (1986). The slides were counterstained with 4' 6-diamidino-2-phenylindole (DAPI) and the metaphases then observed and photographed with an Olympus BX50 epi-fluorescence photomicroscope. The chromosomal morphology was defined based on the arm ratio, as described by Levan et al. (1964), and the chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st) and acrocentric (a), and organized in decreasing order of size in the karyotype.
Results
All of the samples of P. corruscans and P. reticulatum analyzed, i.e., the parental lineages (Figure 1), F1 hybrids ("cachapinta" and "pintachara") (Figure 2), and F2 hybrids and backcrosses (Figure 3), had a diploid number of 56 chromosomes arranged in a karyotype containing 20m+12sm+12st+12a, with no evidence of sex-related chromosomal heteromorphism. C-banding revealed heterochromatic markings in the pericentromeric and terminal portions of some chromosomes in both of the parental and all of the hybrid lineages (Figures 1B,D, 2B,D and 3B,D, respectively).
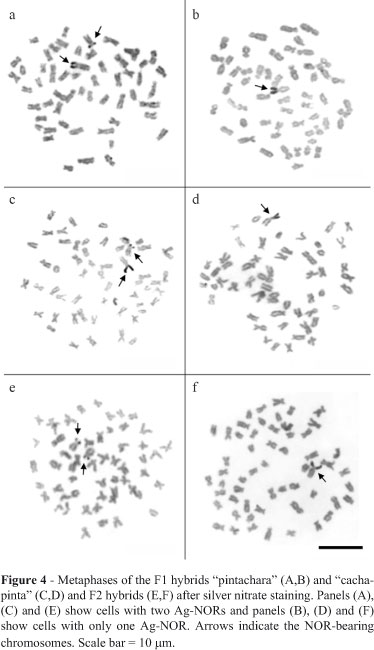
In the parental species, NORs were observed on the short arms of subtelocentric pair 18 (Figure 1A,C) which, in addition to the size polymorphism observed in some metaphases, showed positive C-banding (Figure 1B,D). In the F1, F2 and backcross hybrids, NORs were located on the short arm of two subtelocentric chromosomes of the same size that were polymorphic in some cells (Figures 2A,C and 3A,C) and totally heterochromatic (Figures 2B,D and 3B,D). In addition, silver nitrate staining revealed variations in the distribution of NORs in F1 and F2 hybrids, in which cells with only one active NOR and others with two active NORs were identified (Figure 4 and Table 1).
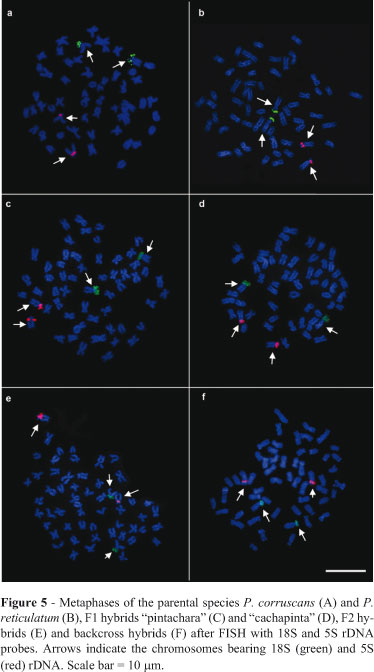
FISH confirmed the location of the 18S ribosomal genes in two subtelocentric chromosomes corresponding to the NOR-bearing chromosomes (Figure 5) in all of the samples studied. The 5S ribosomal genes were detected in the pericentromeric area of the short arm of a subtelocentric pair in P. corruscans and P. reticulatum. In F1 and post-F1 hybrids these genes occurred in the same area of two subtelocentric chromosomes of similar size and morphology (Figure 5).
Discussion
The genus Pseudoplatystoma is characterized by a highly stable number of chromosomes, with all of the species studied so far having 56 chromosomes (Klinkhardt et al., 1995; Arai, 2011). The results described here confirmed this characteristic for P. corruscans and P. reticulatum, both of which had 56 chromosomes arranged in a karyotype composed of 20m+12sm+12st+12a, in agreement with previous reports for these two species (Porto-Foresti et al., 2000; Foresti et al., 2007). However, different karyotypic formulae have been described for P. corruscans (Martins-Santos et al., 1996; Souza et al., 1997; Swarça et al., 2005) and P. reticulatum (Neto et al., 2011), indicating the existence of chromosomal differentiation in these species. Since the differences are subtle and involve few chromosomal pairs, and considering the small size and similar morphology of the chromosomes involved, these discrepancies may be related to difficulties in accurately identifying the chromosomal morphology and in assembling the karyotypes. A similar observation applies to the F1 hybrids analyzed here, both of which maintained the diploid number and chromosomal morphology of their parental lineages.
Closely related species can have the same chromosome number and karyotype that are maintained in the hybrids, as shown for Piaractus mesopotamicus, Colossoma macropomum and their interspecific hybrid "tambacu" (Almeida-Toledo et al., 1988), as well as species of the genus Cichla and their F1 hybrids (Brinn et al., 2004) and hybrids of Oreochromis mossambicus and Oreochromis urolepis hornorum (Zhu et al., 2010).
Hybridization among different fish species can generate not only simple diploid hybrids, with an equal contribution of the parental genomes, but also gynogenetic (containing only the maternal chromosomal group) or androgenetic (containing only the paternal chromosomal group) offspring, as well as haploid, triploid or tetraploid individuals (Toledo-Filho et al., 1994). In the present study, despite the impossibility of identifying the haploid groups of each parental species within the chromosomal group of the interspecific hybrids, these groups can be classified as simple diploid hybrids since molecular studies have shown that these fish inherited specific gene fragments from both of the parental species (Prado et al., 2011).
Fertile hybrids have been described for several fish species (reviewed in Bartley et al., 2001), with the production of post-F1 lineages being more common than in other animals. This reflects the fact that in fish even gametes that have unbalanced chromosomal complements maintain part of their genetic and functional capabilities and can generate viable descendants (Toledo-Filho et al., 1994). However, there have been few cytogenetic studies of post-F1 lineages in fish. One of the few such studies for Neotropical species involved the backcrossing of the hybrid "tambacu" with the parental pacu (Almeida-Toledo et al., 1996), which resulted in a lineage with a cytogenetic pattern identical to that of the parental pacu. The cytogenetic data described here for post-F1 lineages also showed the occurrence of a diploid number and karyotypic constitution identical to the parental species and F1 hybrids.
The C-banding in P. corruscans and P. reticulatum revealed a band pattern characteristic of the family Pimelodidae, with a small amount of constitutive heterochromatin distributed in the terminal and pericentromeric areas of some chromosomes (Fenocchio and Bertollo, 1992; Garcia and Moreira-Filho, 2005). The existence of small differences in the areas of heterochromatin between the parental lineages of cachara and pintado cannot be excluded, although peculiar characteristics of the chromatin of these fish (Fenocchio and Bertollo, 1992) could hinder the detection of discriminatory banding patterns. The finding that the F1 and post-F1 hybrid lineages studied here had the same heterochromatic banding pattern agreed with similar results described by Brinn et al. (2004) for interspecific hybrids of Cichla monoculus and C. temensis.
The NORs in P. corruscans and P. reticulatum were restricted to terminal areas of the chromosomes, as frequently observed in Pimelodidae, and this may represent the ancestral condition for this character in Siluriformes (Oliveira and Gosztonyi, 2000). Differences in the size of the Ag-NOR band among homologous chromosomes are common in Neotropical fish (Foresti et al., 1981) and may be related to variation in NOR activity, to the number of repetitions of rDNA genes, or to other decisive rearrangements involved in determining polymorphism.
The analysis of NORs in the hybrid lineages revealed demarcations in chromosomes of equal size and morphology that hindered identification of the origin of the parental chromosomes. The variation in the number of chromosomes with NORs observed in the F1 and F2 hybrid metaphases was possibly related to differences in the activity of this region since silver nitrate staining is restricted to transcriptionally active NORs (Howell, 1977; Jordan, 1987). Alternatively, such variations may reflect the effect of partial nucleolar dominance in which the NOR of one of the parentals is active while the other one is inactive. This phenomenon is commonly observed in interspecific hybrids and has been described in Drosophila (Durica and Krider, 1978), Xenopus (Reeder and Roan, 1984) and, more recently, in fish (Gold et al., 1991; Hashimoto et al., 2009).
The fact that the backcross hybrids assessed here showed no such variation in their NORs can be explained by the possible "rescue" of the nucleolus lost during backcrossing of the hybrid "cachapinta" with the parental pintado, resulting in active NORs in all of the cells analyzed. In plants of the genus Crepis it was possible "to recover" the repressed nucleolus of interspecific hybrids through appropriate backcrossing, indicating that the nucleolar formation of the chromosome was not altered or lost in the hybrid (Pikaard, 2000).
FISH revealed 18S rDNA clusters in the regions corresponding to NORs in both P. corruscans and P. reticulatum, thus confirming the existence of only one chromosome pair bearing this gene, as is usually the case in the Pimelodidae (Vasconcelos and Martins-Santos, 2000; Swarça et al., 2008; Carvalho et al., 2010). Regardless of the Ag-NOR variation, in situ hybridization in the F1 and F2 hybrids indicated that the 18S ribosomal genes were not lost or altered, as also reported for hybrids of Leporinus (Hashimoto et al., 2009).
Although few chromosomal mapping studies have examined the 5S gene in the Pimelodidae, the evidence indicates the prevalence of only two chromosomes related to these ribosomal sites (Swarça et al., 2009; Carvalho et al., 2010). As shown here, the 5S ribosomal gene was located pericentromerically on one chromosomal pair in P. reticulatum and P. corruscans, as also observed by Swarça et al. (2005) and Neto et al. (2011); the hybrid lineages showed 5S demarcations at the same chromosomal positions as their parental lineages, thus conforming previous observations for this fish group. According to Martins and Galetti Jr (2001), the preferential pericentromeric location of the 5S rDNA sites seen here in the parental and hybrid metaphases may be related to the protection of these sequences from transposition and exchange events that are more frequent in terminal areas. This protection may be an advantageous characteristic in the genomic organization of these organisms.
The techniques used here revealed identical cytogenetic patterns in P. corruscans and P. reticulatum and their maintenance in subsequent F1 and post-F1 hybrid lineages, indicating that chromosomal conservation is an important phenomenon in species of Pimelodidae. A high degree of homology in the chromosomal constitution of the parental species frequently results in correct pairing and segregation during meiosis and the consequent production of viable gametes in interspecific hybrids (Nikoljukin, 1946). This phenomenon could explain the fertility of the F1 hybrids seen here.
Although there may be subtle karyotypic differences between P. corruscans and P. reticulatum, the techniques used here were not sufficiently sensitive to detect such differences among the chromosomal markers of the parental and hybrid lineages. This work represents the first cytogenetic study of F1 and post-F1 hybrids of the pimelodid species P. corruscans and P. reticulatum and the results obtained may be useful in future genetic and evolutionary analyses of the parental species and their hybrid lineages. The findings regarding the reproduction and fertility of the "cachapinta" hybrid represent one of the few studies that have examined the fertility of F1 hybrids and the viability and survival of post-F1 hybrids in Brazilian fish.
Acknowledgments
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Received: April 19, 2011
Accepted: September 21, 2011.
Associate Editor: Marcelo Guerra
License information: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
- Almeida-Toledo LF, Foresti F, Ramos SM, Ormanezi R, Carolsfeld VJS and Toledo Filho SA (1988) Estudos citogenéticos de híbridos entre fêmeas de pacu (Piaractus mesopotamicus) e machos de tambaqui (Colossoma macropomum). Boletim Técnico do CEPTA 1:1-17.
- Almeida-Toledo LF, Bigoni APV, Bernardino G and Toledo-Filho SA (1995) Chromosomal location of NORs and C bands in F1 hybrids of bighead carp and silver carp reared in Brazil. Aquaculture 135:277-284.
- Almeida-Toledo LF, Bernardino G, Oliveira C, Foresti F and Toledo-Filho AS (1996) Gynogenetic fish produced by a backcross involving a male hybrid (female Colossoma macropomum x male Piaractus mesopotamicus) and a female Piaractus mesopotamicus Boletim Técnico do CEPTA 9:31-37.
- Arai R (2011) Fish Karyotypes: A Check List. Springer, Tokyo, 340 pp.
- Bartley DM, Rana K and Immink AJ (2001) The use of inter-specific hybrids in aquaculture and fisheries. Rev Fish Biol Fisheries 10:325-337.
- Brinn MNA, Porto JIR and Feldberg E (2004) Karyological evidence for interspecific hybridization between Cichla monoculus and C. temensis (Perciformes, Cichlidae) in the Amazon. Hereditas 141:252-257.
- Buitrago-Suárez UA and Burr BM (2007) Taxonomy of the catfish genus Pseudoplatystoma Blecker (Siluriformes, Pimelodidae) with recognition of eight species. Zootaxa 1512:1-38.
- Campos JL (2010) O cultivo do pintado (Pseudoplatystoma corruscans, Spix; Agassiz, 1829), outras espécies do gênero Pseudoplatystoma e seus híbridos. In: Baldisserotto B and Gomes LC (eds) Espécies Nativas para Piscicultura no Brasil. UFMS, Campo Grande, pp 335-361.
- Carvalho RA, Laudicina A, Giuliano-Caetano L, Martins-Santos IC and Dias AL (2010) Cytogenetic analysis of the 18S, 5S rDNA and B chromosome of Iheringichthys labrosus (Lütken, 1874) (Siluriformes, Pimelodidae). Braz J Biol 70:631-636.
- Durica DS and Krider HM (1978) Studies on the ribosomal RNA cistrons in Drosophila hybrids. II: Heterochromatic regions mediating nucleolar dominance. Genetics 89:37-64.
- Fenocchio AS and Bertollo LAC (1992) Karyotype similarities among Pimelodidae (Pisces, Siluriformes) from the Brazilian Amazon region. Cytobios 69:41-46.
- Ferraris Jr CJ (2007) Checklist of catfishes, recent and fossil (Osteichthyes, Siluriformes), and catalogue of siluriform primary types. Zootaxa 1418:329-345.
- Foresti F (2008) A brief history of fish genetics in Brazil. Genet Mol Biol 31(suppl):385-388.
- Foresti F, Almeida-Toledo LF and Toledo-Filho SA (1981) Polymorphic nature of nucleolus organizer regions in fishes. Cytogenet Cell Genet 31:137-144.
- Foresti F, Senhorini JA, Almeida-Toledo LF, Oliveira C, Porto-Foresti F, Silva MFZD and Calcagnotto D (2007) Análise citogenética e genético-molecular das populações de Piaractus mesopotamicus, Brycon hilarii, Pseudoplatystoma corruscans, Prochilodus lineatus e Leporinus macrocephalus dos rios Miranda, Aquidauana, Paraguai (MS), Cuiabá/Manso e do Parque Nacional do Pantanal Mato-Grossense (MT). In: Ministério do Meio Ambiente (ed). Pesquisas Patológicas e Genéticas em Recursos Pesqueiros da Bacia do Alto Paraguai. IBAMA, Brasília, pp 128-164.
- Garcia C and Moreira-Filho O (2005) Cytogenetical analyses in three fish species of the genus Pimelodus (Siluriformes, Pimelodidae) from rio São Francisco: Considerations about the karyotypical evolution in the genus. Neotrop Ichthyol 3:285-290.
- Godinho HP (2007) Estratégias reprodutivas de peixes aplicadas à aqüicultura: Bases para o desenvolvimento de tecnologias de produção. Rev Bras Reprod Anim 31:351-360.
- Gold JR, Li Y, Schmidt TR and Tave D (1991) Nucleolar dominance in interspecific hybrids of cyprinid fishes. Cytobios 65:139-147.
- Hashimoto DT, Laudicina A, Bortolozzi J, Foresti F and Porto-Foresti F (2009) Chromosomal features of nucleolar dominance in hybrids between the Neotropical fish Leporinus macrocephalus and Leporinus elongatus (Characiformes, Anostomidae). Genetica 137:135-140.
- Howell WM (1977) Visualization of ribosomal gene activity: Silver stains proteins associated with rRNA transcribed from oocyte chromosomes. Chromosoma 62:361-367.
- Howell WM and Black DA (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 36:1014-1015.
- Jordan G (1987) At the heart of the nucleolus. Nature 329:489-490.
- Klinkhardt M, Tesche M and Greven H (1995) Database on fish chromosomes. Westarp Wissenschaften, Hohenwarsleben, 237 pp.
- Levan A, Fredga K and Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201-220.
- Lozano R, Rejon CR and Rejon MR (1988) A method for increasing the number of mitoses avaliable for cytogenetic analysis in rainbow trout. Stain Technol 66:335-338.
- Lundberg JG and Littmann MW (2003) Family Pimelodidae (Long-whiskered catfishes). In: Reis R, Kullander SO and Ferraris Jr CJ (eds) Check List of the Freshwater Fishes of South and Central America. Edipucrs, Porto Alegre, pp 432-455.
- Martins C and Galetti Jr PM (2001) Two 5S rDNA arrays in Neotropical fish species: Is it a general rule for fishes? Genetica 111:439-446.
- Martins-Santos IC, Julio-Jr HF and Burin I (1996) Karyotypic studies of four species of the Sorubiminae subfamily (Pisces, Siluriformes). Caryologia 49:73-80.
- Neto AM, Silva M, Matoso DA, Vicari MR, Almeida MC, Collares-Pereira MJ and Artoni RF (2011) Karyotype variability in neotropical catfishes of the family Pimelodidae (Teleostei, Siluriformes). Neotrop Ichthyol 9:97-105.
- Nikoljukin NI (1946) Experimental progeny obtained from intergeneric hybrids of Cyprinidae. Comp Rend (Dokl) Acad USSR 51:737-740.
- Oliveira C and Gosztonyi AE (2000) A cytogenetic study of Diplomystes mesembrinus (Teleostei, Siluriformes, Diplomystidae) with a discussion on chromosome evolution in Siluriforms. Caryologia 53:31-37.
- Oliveira C, Almeida-Toledo LF, Foresti F and Toledo-Filho SA (1988) Supernumerary chromosomes, Robertsonian rearrangements and multiple NORs in Corydoras aeneus (Pisces, Siluriformes, Callichthyidae). Caryologia 41:227-236.
- Oliveira C, Foresti F and Hilsdorf AWS (2009) Genetics of Neotropical fish: From chromosomes to populations. Fish Physiol Biochem 35:81-100.
- Pendás AM, Morán P, Freije JP and Garcia-Vásquez E (1994) Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenet Cell Genet 67:31-36.
- Pikaard CS (2000) The epigenetics of nucleolar dominance. Trends Genet 16:495-499.
- Pinkel D, Straume T and Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934-2938.
- Porto-Foresti F and Foresti F (2004) Genética e biotecnologia em piscicultura: Usos na produção, manejo e conservação dos estoques de peixes. In: Cyrino JEP, Urbinati EC, Fracalossi DM and Castagnolli N (eds) Tópicos Especiais em Piscicultura de Água Doce Tropical Intensiva. TecArt, São Paulo, pp 195-215.
- Porto-Foresti F, Andreata AA, Oliveira C and Foresti F (2000) The karyotype of Pseudoplatystoma fasciatum (Teleostei, Siluriformes) from the Rio Paraguay basin. Chromosome Sci 4:99-102.
- Porto-Foresti F, Hashimoto DT, Alves AL, Almeida RBC, Senhorini JA, Bortolozzi J and Foresti F (2008) Cytogenetic markers as diagnoses in the identification of the hybrid between piauçu (Leporinus macrocephalus) and piapara (Leporinus elongatus). Genet Mol Biol 31(suppl):195-202.
- Prado FD, Hashimoto DT, Mendonça FF, Senhorini JA, Foresti F and Porto-Foresti F (2011) Molecular identification of hybrids between Neotropical catfish species Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum Aquacult Res (Epub).
- Reeder RH and Roan JG (1984) The mechanism of nucleolar dominance in Xenopus hybrids. Cell 38:39-44.
- Souza AB, Fonseca CG, Ribeiro LP and Pinheiro LEL (1997) Análise cromossômica do surubim Pseudoplatystoma coruscans das bacias dos rios São Francisco e Paraguai. In: Miranda MOT (ed) Surubim: Coleção Meio Ambiente, Série Estudos da Pesca. IBAMA, Belo Horizonte, pp 57-68.
- Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304-306.
- Swarça AC, Fenocchio AS, Cestari MM and Dias LA (2005) Karyotype divergence among populations of giant catfish Pseudoplatystoma corruscans (Teleostei, Pimelodidae) indicates higher species diversity. Ichthyol Explor Freshwaters 16:325-330.
- Swarça AC, Fenocchio AS, Cestari MM and Dias LA (2008) Analyses of the structure of NORs in two species of South American Sorubiminae fishes (Siluriformes) by means of several cytogenetic techniques. Folia Biol (Kraków) 56:31-35.
- Swarça AC, Fenocchio AC, Cestari MM and Dias AL (2009) Localization and characterization of the 5S rDNA bearing chromosome in two Steindachneridion species by means of different dytogenetic techniques. Cytologia 74:323-327.
- Toledo-Filho SA, Almeida-Toledo LF, Foresti F, Bernardino G and Calcagnotto D (1994) Monitoramento e Conservação Genética em Projeto de Hibridação entre Pacu e Tambaqui. Cadernos de Ictiogenética 2. CCS/USP, São Paulo, 48 pp.
- Toledo-Filho SA, Almeida-Toledo LF, Foresti F, Calcagnotto D, Santos SBAF and Bernardino G (1998) Programas Genéticos de Seleção, Hibridação e Endocruzamento Aplicados à Piscicultura. Cadernos de Ictiogenética 4. CCS/USP, São Paulo, 56 pp.
- Vasconcelos C and Martins-Santos IC (2000) Chromosome polymorphism in species of the Pimelodidae family (Pisces, Siluriformes). Hereditas 132:103-109.
- White TJ, Bruns T, Lee S and Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ and White TJ (eds) PCR Protocols - A Guide to Methods and Applications. Academic Press, San Diego, pp 315-322.
- Zhu HP, Lu MX, Gao FY, Huang ZH, Yang LP and Gui JF (2010) Chromosomal localization of rDNA genes and genomic organization of 5S rDNA in Oreochromis mossambicus, O. urolepis hornorum and their hybrid. J Genet 89:163-171.
Publication Dates
-
Publication in this collection
02 Feb 2012 -
Date of issue
2012
History
-
Received
19 Apr 2011 -
Accepted
21 Sept 2011