Abstract
Snake liver has been implicated in the adaptation of snakes to a variety of habitats. However, to date, there has been no systematic analysis of snake liver proteins. In this study, we undertook a proteomic analysis of liver from the colubrid snake Elaphe taeniura using a combination of two-dimensional electrophoresis (2-DE) and matrix-assisted laser desorption/ionization time of flightmass spectrometry (MALDI-TOF MS). We also constructed a local protein sequence database based on transcriptome sequencing to facilitate protein identification. Of the 268 protein spots revealed by 2-DE 109 gave positive MS signals, 84 of which were identified by searching the NCBInr, Swiss-Prot and local databases. The other 25 protein spots could not be identified, possibly because their transcripts were not be stable enough to be detected by transcriptome sequencing. GO analysis showed that most proteins may be involved in binding, catalysis, cellular processes and metabolic processes. Forty-two of the liver proteins identified were found in other reptiles and in amphibians. The findings of this study provide a good reference map of snake liver proteins that will be useful in molecular investigations of snake physiology and adaptation.
2D electrophoresis; Elaphe taeniura; MALDI-TOF mass spectrometry; protein
GENOMICS AND BIOINFORMATICS
RESEARCH ARTICLE
Proteomic profiling of liver from Elaphe taeniura, a common snake in eastern and southeastern Asia
Liang ChenI; Hengchuan XiaIII; Yiting WangII; Keping ChenIII; Lvgao QinIII; Bin WangIII; Qin YaoIII; Jun LiIV; Yuanqing HeIII; Ermi ZhaoI,V
IKey Laboratory of Bio-resources and Eco-environment, College of Life Sciences, Sichuan University, Chengdu, Sichuan Province, China
IICollege of Athletic Sports, Yangzhou University, Yangzhou, Jiangsu Province, China
IIIInstitute of Life Sciences, Jiangsu University, Zhenjiang, Jiangsu Province, China
IVCenter for Physics and Chemistry, Jiangsu University, Zhenjiang, Jiangsu Province, China
VChengdu Institute of Biology, the Chinese Academy of Sciences, Chengdu, Sichuan Province, China
Send correspondence to Send correspondence to Ermi Zhao Key Laboratory of Bio-Resources and Eco-environment, College of Life Sciences, Sichuan University, No.24 South Section 1, Yihuan Road, 610064 Chengdu, Sichuan Province, China. E-mail: ermizhao@gmail.com
ABSTRACT
Snake liver has been implicated in the adaptation of snakes to a variety of habitats. However, to date, there has been no systematic analysis of snake liver proteins. In this study, we undertook a proteomic analysis of liver from the colubrid snake Elaphe taeniura using a combination of two-dimensional electrophoresis (2-DE) and matrix-assisted laser desorption/ionization time of flightmass spectrometry (MALDI-TOF MS). We also constructed a local protein sequence database based on transcriptome sequencing to facilitate protein identification. Of the 268 protein spots revealed by 2-DE 109 gave positive MS signals, 84 of which were identified by searching the NCBInr, Swiss-Prot and local databases. The other 25 protein spots could not be identified, possibly because their transcripts were not be stable enough to be detected by transcriptome sequencing. GO analysis showed that most proteins may be involved in binding, catalysis, cellular processes and metabolic processes. Forty-two of the liver proteins identified were found in other reptiles and in amphibians. The findings of this study provide a good reference map of snake liver proteins that will be useful in molecular investigations of snake physiology and adaptation.
Keywords: 2D electrophoresis, Elaphe taeniura, MALDI-TOF mass spectrometry, protein.
Introduction
Proteomics has been widely used for the global analysis of protein profiles and has provided insights into mechanisms related to development, physiology and pathology (Yanes et al., 2007). Proteomic studies have been reported for various organisms, such asbacteria, mouse, rat, guinea pig (Cavia porcellus), worms (Nematoda), Leguminosae (Medicago sativa Linn), rice (Oryza sativa) and humans (Klose, 1975; O'Farrell, 1975; Scheele, 1975; Emmert-Buck et al., 2000; Kaji et al., 2000; Ahram et al., 2002; Li et al., 2003), as well as amphibians, e.g., the African clawed frog Xenopus laevis (Jelaso et al., 2005; Gillardin et al., 2009), and snakes (Campbell and Campbell,2001). Although proteomic analyses of venoms from more than 55 snake genera have been reported (Fry et al., 2003; Serrano et al., 2005; Menezes et al., 2006; Yanes et al., 2007; Fox and Serrano, 2008), to date there has been no proteomic analysis of snake tissues.
Snakes are highly adapted to a variety of terrestrial, arboreal, semiaquatic and marine habitats (Conant and Collins,1991). This adaptability has been attributed to the development of a special metabolic system in snakes. The liver plays a major role in metabolism, including glycogen storage, red blood cell degradation, plasma protein synthesis, hormone production and detoxification (Dardevet et al., 2006; Reddy and Rao, 2006), and also contains disease-associated proteins (Zeindl-Eberhart et al., 2001; Yokoyama et al., 2004). Recent studies have shown that snake liver has a strong metabolic capacity (Ladyman et al., 2003; Starck et al., 2004) that may have an important role ensuring survival in nature. Previous studies of snake liver have focused mainly on plasma proteins (Chiu and Lam,1994), as well as histological and biochemical analyses (Chiu and Wong, 1974; Wong and Chiu, 1979; Chang and Zheng, 2003); there have been no molecular biology or proteomic analyses.
In this report, we describe the first proteomic study of snake liver using 2-DE and MALDI-TOF MS. The source of liver was Elaphe taeniura (Cope, 1861; Pike et al., 2008), a colubrid snake with a wide distribution in China, Russian, Japan, India and southeastern Asia. Since there are no reports on snake genomes, we also constructed a local database of predicted protein sequences based on transcriptome sequencing data; this database significantly facilitated protein identification. A reference map of the E. taeniura liver protein profile was successfully constructed and 84 of 268 protein spots were identified.
Materials and Methods
Preparation
Adult E. taeniura were provided by Nanjing Jinlin Snake Industry Co. Ltd. (Jiangsu province, China). After anesthesia by an intraperitoneal injection of an overdose (45 mg/kg) of sodium pentobarbital, we dissected the body of snakes. Samples of the liver tissue were immediately frozen in liquid nitrogen and stored at -70 ºC until use. All of the procedures conformed to ethical standards for the treatment of animals and were designed to minimize animal suffering. The protocol was approved by the ethics committee of the Laboratory for Animal Research Center at Jiangsu University, China. All reagents were from Sigma unless stated otherwise.
Protein sample preparation
Hepatic tissue (0.15 g) was homogenized with a precooled mortar and pestle in 1.5 mL of lysis buffer containing 7 M urea, 2 M thiourea and 4% Chaps followed by sonication (twice) for 10 s on ice. The homogenate was transferred to an Eppendorf tube and centrifuged (10,000 g, 20 min, 4 ºC). Protein samples were treated with a 2-DE clean-up kit (GE Healthcare), vacuum-dried and dissolved in 120 µL of buffer A containing 7 M urea, 2 M thiourea, 4% Chaps, 0.2% Bio-Lyte pH 3-10 and 65 mM dithiotreitol (DTT). The protein concentration was determined using the Bradford assay with bovine serum albumin (BSA) as standard. The protein samples were stored at -20 ºC for later use.
Two-dimensional electrophoresis
Isoelectric focusing (IEF) was done using an IPGphor focusing system (Bio-Rad) according to the manufacturer's instructions. IPG strips (linear pH 3-10 gradient; 17 cm) were run at 20 ºC. The protein sample (1.4 mg) was first rehydrated in buffer using active rehydration (13 h with 50 V) in a total volume of 400 µL after which IEF was done with a voltage gradient of 250 V (0.5 h), 1,000 V (1 h) and 10,000 V (5 h), and then continued for a total of 60 kVh at 10 kV. The focused strip was equilibrated for 15 min with equilibration solution (6 M urea, 0.375 M Tris-HCl, pH 8.8, 20% (v/v) glycerol, 2% (w/v) SDS and 0.002% (w/v) bromophenol blue) containing 2% (w/v) DTT and for another 15 min with same solution containing 2.5% (w/v) iodoacetamide. SDS-PAGE was done using 12% gels at 30 mA (constant) until the dye front reached the bottom of the gel. The gel was stained with 0.1% coomassie brilliant blue G-250 (Bio-SafeTM, Bio-Rad) and photographed with a digital single lens reflex (Nikon D5000) and standard lens (AF 50 mm f/1.4D).
In situ digestion
268 protein spots in the 2D gel that were detected by coomassie brilliant blue staining were manually excised, transferred to Eppendorf tubes and then destained, reduced, alkylated and digested with trypsin. Briefly, the gel slices were immersed in double-distilled water, sonicated (twice) for 10 min, washed with 100 mM NH4HCO3/acetonitrile (1:1, v/v), dehydrated in acetonitrile and dried in a Speedvac vacuum concentrator (Eppendorf AG, Hamburg, Germany). The protein slices were then reduced with 10 mM DTT in 100 mM NH4HCO3 for1hat56ºC and then incubated with 40 mM iodoacetamide in 100 mM NH4HCO3 for 45 min at room temperature. The protein spots were washed sequentially with 25 mM ammonium bicarbonate, 50% acetonitrile and 100% acetonitrile, and then incubated overnight in 25 mM NH4HCO3 containing 20 µg of trypsin/mL at 37 ºC (Liang et al., 2007; Qin et al., 2009).
Mass spectrometry analysis
After digestion, the protein peptides were extracted twice in 0.5% trifluoroacetic acid (TFA) and 2.5% TFA/50% acetonitrile. A one microliter sample was then spotted onto an MTP Anchor Chip board (Bruker) and analyzed with a MALDI-TOF mass spectrometer. Peptide mass fingerprints of 1,000-4,000 Da were obtained. Standard peptides were used as external standards. The peak value of the trypsin peptide and matrix were used as internal parameters (Kim et al., 2007).
Liver transcriptome of E. taeniura
The liver transcriptome of E. taeniura was sequenced by Hanyu Biological Co., Ltd. (Shanghai, China). Briefly, liver RNA was extracted and mRNA was purified by standard procedures followed by cDNA synthesis. RNA was fragmented by incubation at 95 ºC for 5 min and annealed with biotinylated random primers that contained a Solexa adapter sequence. The RNA fragments were then captured with streptavidin coupled to biotinylated random primers. Finally, a double-strand Solexa library was produced by PCR amplification, with clean data being obtained from the raw data using the FASTX software package. De novo assembling was then done using the Velvet and Oases software packages.
Bioinformatics analysis
Open reading frames were identified by using an in-house program based on 'GetORF' from EMBOSS (Rice et al., 2000). Gene annotation was done by BLASTP searching against the Swiss-Prot and GenBank databases with an E value cutoff of 1e-3. The predicted proteins were used to construct a local MASCOT protein database and to analyze the peptide finger prints of snake liver proteins.
To identify the proteins, the MS fingerprints were screened against the NCBInr and Swiss-Prot sequence databases with the search engine MASCOT. Unidentified proteins were searched in a local database constructed specifically for this purpose. Fixed modification was set to be carbamidomethyl (C) and variable modification was set to be oxidation (M). The mass tolerance was set as ± 0.2 to ± 0.8 Da. The species were set as Chordata and a CI score > 62 was considered to be a positive match.
The resulting protein sequences were aligned with Interpro Scan to obtain the gene ontology (GO) identifications and the collected information was then analyzed by WEGO (Ye et al., 2006).
Configuration and installation of local database
The Configuration Editor/Database Maintenance of the MASCOT homepage was used for the custom database configuration. In this process, the translated protein sequence in sequence directory file is parsed and compressed by the MASCOT program, producing homonyms a00, i00, s00 and a statistics database retrieval file in which the MS data perform a retrieval step. Subsequently, the new data-base name (cldata) was entered in the EST sequence file path in the configuration page, options and parsing rules were defined and applied to complete a new database definition. Details on the settings can be provided by the authors on request.
Results
Protein profile of E. taeniura liver
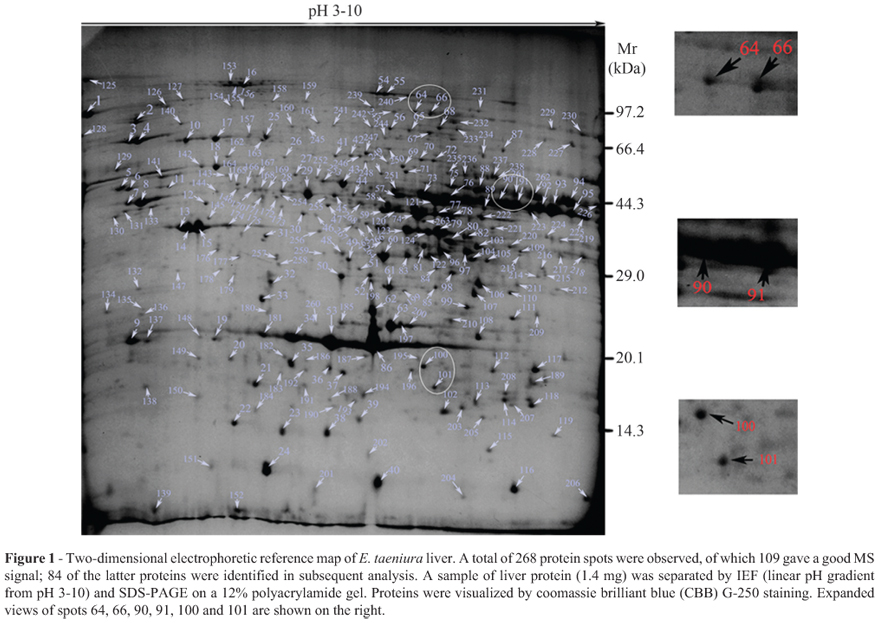
Figure 1 shows a reference 2-DE map of E. taeniura liver in which 268 spots were detected. The pIs of 196 protein spots ranged from 5 to 9 (73.1% of spots); 21 spots had a pI < 4 (7.83% of spots) and 26 had a pI > 9 (9.7% of spots). The molecular masses of most proteins were between 29 kDa and 97.2 kDa. 109 protein spots had a good MS signal, 84 of which were identified. Of these 84 proteins, 54 were represented in the NCBInr and Swiss-Prot databases for Chordata and 30 were in the local database. Table 1 summarizes the data for these proteins.
Characteristics of the transcriptome assembly
DNA sequencing by Illumina HiSeq 2000 resulted in a cDNA library containing 29,614,448 reads and 2,754,143,664 bases. The number of paired-end reads was 14,807,224, with an average length of 93 bp. A total of 88,907 contigs > 100 bp in size (which reflected the quality of the data) was assembled using Velvet and Oases softwares, and 23,285 predicted proteins had clear biological annotations.
Protein identification and GO analysis
Of the 84 identified proteins, 42 were matched to amphibian and reptile protein sequence databases. Interestingly, the amphibian and reptile databases contained no information for the remaining 42 proteins, although there were matches in databases of other species. The presence of these protein spots in the 2D gel indicates that many snake proteins remain to be identified. For instance, protein spot 74 was identified as tau-crystallin, a homologue of which has been found mainly in crocodiles. Figure 2 shows the peptide fingerprint and peptide match for spot 74 obtained with the MASCOT database. GO analysis showed that these proteins are mainly involved in binding, catalysis, cellular processes and metabolic processes (Figure 3).
Discussion
In this study, we successfully constructed a reference map for the liver protein profile of the colubrid snake E. taeniura. To date, liver proteomic analyses have been reported for mammals and fish (Karim et al., 2011; Molette et al., 2012). As far as we know, the present report is the first to describe the protein profile for snake liver. Preliminary experiments showed that common protein extraction methods such as alcohol phenyl, TCA/acetone, schizolysis were not applicable to snake liver (unpublished results). Instead, a combination of a 2D clean-up kit with the method described here significantly improved the resolution, and 268 protein spots were detected in 2D gels after staining with Coomassie blue. Further optimization of the protein extraction and visualization method could be helpful in identifying even more proteins.
As the snake genome has not yet been described and many proteins could not be identified by searching the NCBInr and Swiss-Prot databases, we constructed a local database based on transcriptome sequencing data. As a result, 84 of the 109 protein spots with high MS scores were identified. Interestingly, the remaining 25 proteins could not be identified although they were present in the 2D gels (Figure 1); these proteins may be exclusive to snakes. In addition, some protein spots, e.g., spots 64 and 66, and spots 90 and 91, were found to be the same proteins, although they migrated at different positions in 2-DE. This is a common finding in proteomic studies, as some proteins vary in their post-translational modifications (Meri and Baumann, 2001; Bretteville et al., 2009; Merkley et al., 2009). For example, protein spots 64 and 66 were identified as aconitate hydratase, which is predicted to have 11 phosphorylation sites. Similarly, protein spots 100 and 101 corresponded to dimeric 4-hydroxyphenylpyruvate dioxygenase (Lindblad et al., 1970; Ruetschi et al., 1997), the dimeric nature of which could contribute to variation in the migration pattern in 2-DE. GO analysis showed that most of the proteins identified were involved in catalytic activity, metabolic processes and binding activity, a finding consistent with liver proteomic data for other species (Mi et al., 2007; Wei et al., 2007).
Heat shock proteins (Hsps) Hsp60, Hsp70 and Hsp90 (spots 18, 25 and 5, respectively, in Figure 1) were highly expressed in liver tissue of E. taeniura. Hsps are a family of proteins induced by diverse stress factors, such as an increase in temperature, infection, inflammation, starvation, hypoxia and water deprivation. These proteins play important roles in protein folding, translocation and the assembly of intracellular proteins that may protect against environmental stress (Gray et al., 1999; Nollen and Morimoto, 2002; Sun et al., 2008; Di Domenico et al., 2010). In the face of increasing habitat destruction, many animals, including reptiles, are facing severe environmental challenges that require physiological adaptations. The elevated expression of Hsps in snake liver may help to protect this species from such stress and could contribute to the ability of snakes to adapt to different environments.
Some of the proteins identified here, e.g., tau-crystallin (spot 74) and a-enolase (spots 59, 77 and 78), have also been found in amphibians and reptiles. Tau-crystallin is a taxon-restricted crystallin found in the eye lenses of reptiles and a few avian species but is presumably absent in mammals (Mishra et al., 2002). The level of tau-crystallin expression in the lens varies among species, e.g., in crocodiles it is the least abundant crystallin and is present in trace amounts (Williams et al., 1985; Mishra et al., 2002). Interestingly, tau-crystallin had a relatively high expression in E. taeniura liver.
Acknowledgments
This work was supported by Natural Science Fundation for Universities of Jiangsu Province (No. 10KJB180001), Start-Up Research Funding of Jiangsu University for Distinguished Scholars (No.09JDG005), and Postdoctoral daily finance of Jiangsu University (1143002139). We thank professors Xiaoyong Liu and Haijun Liu for their scientific advice and technical expertise.
Internet Resources
Received: November 14, 2012; Accepted: May 11, 2013.
Associate Editor: Ana Tereza R. Vasconcelos
License information: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
- Ahram M, Best CJ, Flaig MJ, Gillespie JW, Leiva IM, Chuaqui RF, Zhou G, Shu H, Duray PH, Linehan WM, et al. (2002) Proteomic analysis of human prostate cancer. Mol Carcinog 33:9-15.
- Bretteville A, Ando K, Ghestem A, Loyens A, Begard S, Beauvillain JC, Sergeant N, Hamdane M and Buee L (2009) Two-dimensional electrophoresis of tau mutants reveals specific phosphorylation pattern likely linked to early tau conformational changes. PloS One 4:e4843.
- Campbell KR and Campbell TS (2001) The accumulation and effects of environmental contaminants on snakes: A review. Environ Monit Assess 70:253-301.
- Chang C and Zheng R (2003) Effects of ultraviolet B on epidermal morphology, shedding, lipid peroxide, and antioxidant enzymes in Cope's rat snake (Elaphe taeniura). J Photochem Photobiol B 72:79-85.
- Chiu K and Lam K (1994) Plasma T3 and T4 levels in a snake, Elaphe taeniura Comp Biochem Physiol A 107:107-112.
- Chiu KW and Wong CC (1974) Testicular cycles of the striped racer, Elaphe taeniura Gen Comp Endocrinol 23:242-244.
- Conant R and Collins J (1991) A Field Guide to Amphibians and Reptiles of Eastern and Central North America. Houghton Mifflin Co, Boston, 450 pp.
- Cope ED (1861) Catalogue of the Colubridae in the Museum of the Academy of Natural Sciences of Philadelphia. Part 3. Proc Acad Nat Sci Philadelphia 12:553-566.
- Dardevet D, Moore MC, Remond D, Everett-Grueter CA and Cherrington AD (2006) Regulation of hepatic metabolism by enteral delivery of nutrients. Nutr Res Rev 19:161-173.
- Di Domenico F, Sultana R, Tiu GF, Scheff NN, Perluigi M, Cini C and Butterfield DA (2010) Protein levels of heat shock proteins 27, 32, 60, 70, 90 and thioredoxin-1 in amnestic mild cognitive impairment: An investigation on the role of cellular stress response in the progression of Alzheimer disease. Brain Res 1333:72-81.
- Emmert-Buck MR, Gillespie JW, Paweletz CP, Ornstein DK, Basrur V, Appella E, Wang QH, Huang J, Hu N, Taylor P, et al. (2000) An approach to proteomic analysis of human tumors. Mol Carcinog 27:158-165.
- Fox JW and Serrano SM (2008) Exploring snake venom proteomes: Multifaceted analyses for complex toxin mixtures. Proteomics 8:909-920.
- Fry BG, Wuster W, Ryan Ramjan SF, Jackson T, Martelli P and Kini RM (2003) Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Commun Mass Spectrom 17:2047-2062.
- Gillardin V, Silvestre F, Dieu M, Delaive E, Raes M, Thome JP and Kestemont P (2009) Protein expression profiling in the African clawed frog Xenopus laevis tadpoles exposed to the polychlorinated biphenyl mixture aroclor 1254. Mol Cell Proteomics 8:596-611.
- Gray CC, Amrani M and Yacoub MH (1999) Heat stress proteins and myocardial protection: Experimental model or potential clinical tool? Int J Biochem Cell Biol 31:559-573.
- Jelaso AM, Delong C, Means J and Ide CF (2005) Dietary exposure to Aroclor 1254 alters gene expression in Xenopus laevis frogs. Environ Res 98:64-72.
- Kaji H, Tsuji T, Mawuenyega KG, Wakamiya A, Taoka M and Isobe T (2000) Profiling of Caenorhabditis elegans proteins using two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-time of flight-mass spectrometry. Electrophoresis 21:1755-1765.
- Karim M, Puiseux-Dao S and Edery M (2011) Toxins and stress in fish: Proteomic analyses and response network. Toxicon 57:959-969.
- Kim Y, Nandakumar MP and Marten MR (2007) Proteome map of Aspergillus nidulans during osmoadaptation. Fungal Genet Biol 44:886-895.
- Klose J (1975) Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik 26:231-243.
- Ladyman M, Bonnet X, Lourdais O, Bradshaw D and Naulleau G (2003) Gestation, thermoregulation, and metabolism in a viviparous snake, Vipera aspis: Evidence for fecundityindependent costs. Physiol Biochem Zool 76:497-510.
- Li WW, Quinn GB, Alexandrov NN, Bourne PE and Shindyalov IN (2003) A comparative proteomics resource: Proteins of Arabidopsis thaliana Genome Biol 4:R51.
- Liang Y, Chen H, Tang M and Shen S (2007) Proteome analysis of an ectomycorrhizal fungus Boletus edulis under salt shock. Mycol Res 111:939-946.
- Lindblad B, Lindstedt S, Olander B and Omfeldt M (1970) Purification ofp-hydroxyphenylpyruvate hydroxylase from human liver. Acta Chem Scand 25:329-330.
- Menezes MC, Furtado MF, Travaglia-Cardoso SR, Camargo AC and Serrano SM (2006) Sex-based individual variation of snake venom proteome among eighteen Bothrops jararaca siblings. Toxicon 47:304-312.
- Meri S and Baumann M (2001) Proteomics: Posttranslational modifications, immune responses and current analytical tools. Biomol Eng 18:213-220.
- Merkley MA, Hildebrandt E, Podolsky RH, Arnouk H, Ferris DG, Dynan WS and Stoppler H (2009) Large-scale analysis of protein expression changes in human keratinocytes immortalized by human papilloma virus type 16 E6 and E7 oncogenes. Proteome Sci 7:29.
- Mi J, Garcia-Arcos I, Alvarez R and Cristobal S (2007) Age-related subproteomic analysis of mouse liver and kidney peroxisomes. Proteome Sci 5:19.
- Mishra AK, Chandrashekhar R, Aggarwal RK and Sharma Y (2002) Crocodilian tau-crystallin: Overexpression, purification, and characterization. Protein Expr Purif 25:59-64.
- Molette C, Théron L, Marty-Gasset N, Fernandez X and Rémignon H (2012) Current advances in proteomic analysis of (fatty) liver. J Proteomics 75:4290-4295.
- Nollen EA and Morimoto RI (2002) Chaperoning signaling pathways: Molecular chaperones as stress-sensing 'heat shock' proteins. J Cell Sci 115:2809-2816.
- O'Farrell pH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007-4021.
- Pike DA, Pizzatto L, Pike BA and Shine R (2008) Estimating survival rates of uncatchable animals: The myth of high juvenile mortality in reptiles. Ecology 89:607-611.
- Qin L, Liu X, Li J, Chen H, Yao Q, Yang Z, Wang L and Chen K (2009) Protein profile of Nomuraea rileyi spore isolated from infected silkworm. Curr Microbiol 58:578-585.
- Reddy JK and Rao MS (2006) Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol 290:G852-858.
- Rice P, Longden I and Bleasby A (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16:276-277.
- Ruetschi U, Rymo L and Lindstedt S (1997) Human 4-hydroxyphenylpyruvate dioxygenase gene (HPD). Genomics 44:292-299.
- Scheele GA (1975) Two-dimensional gel analysis of soluble proteins. Characterization of guinea pig exocrine pancreatic proteins. J Biol Chem 250:5375-5385.
- Serrano SM, Shannon JD, Wang D, Camargo AC and Fox JW (2005) A multifaceted analysis of viperid snake venoms by two-dimensional gel electrophoresis: An approach to understanding venom proteomics. Proteomics 5:501-510.
- Starck JM, Moser P, Werner RA and Linke P (2004) Pythons metabolize prey to fuel the response to feeding. Proc Biol Sci 271:903-908.
- Sun B, Wang JH, Lv YY, Zhu SS, Yang J and Ma JZ (2008) Proteomic adaptation to chronic high intensity swimming training in the rat heart. Comp Biochem Physiol Part D Genomics Proteomics 3:108-117.
- Wei Y, Wang J, Zhang X, Xu M and Dai J (2007) Identification and characterization of expressed sequence tags from the liver of rare minnow (Gobiocypris rarus). Comp Biochem Physiol D 2:356-362.
- Williams LA, Ding L, Horwitz J and Piatigorsky J (1985) tau-Crystallin from the turtle lens: Purification and partial characterization. Exp Eye Res 40:741-749.
- Wong C and Chiu K (1979) The snake thyroid gland V. Effects of hypophysectomy on iodine metabolism in the striped racer, Elaphe taeniura J Comp Physiol B 131:297-302.
- Yanes O, Aviles FX, Wenzel R, Nazabal A, Zenobi R and Calvete JJ (2007) Proteomic profiling of a snake venom using high mass detection MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom 18:600-606.
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R and Bolund L (2006) WEGO: A web tool for plotting GO annotations. Nucleic Acids Res 34:W293-297.
- Yokoyama Y, Kuramitsu Y, Takashima M, Iizuka N, Toda T, Terai S, Sakaida I, Oka M, Nakamura K and Okita K (2004) Proteomic profiling of proteins decreased in hepatocellular carcinoma from patients infected with hepatitis C virus. Proteomics 4:2111-2116.
- Zeindl-Eberhart E, Klugbauer S, Dimitrijevic N, Jungblut PR, Lamer S and Rabes HM (2001) Proteome analysis of rat hepatomas: Carcinogen-dependent tumor-associated protein variants. Electrophoresis 22:3009-3018.
- Interpro Scan, http://www.ebi.ac.uk/Tools/pfa/iprscan/ (accessed August 28, 2012).
» link - WEGO, http://wego.genomics.org.cn/cgi-bin/wego/index.pl (accessed August 30, 2012).
» link
Send correspondence to
Publication Dates
-
Publication in this collection
23 Aug 2013 -
Date of issue
2013
History
-
Received
14 Nov 2012 -
Accepted
11 May 2013