Abstract
Genes that contain small open reading frames (smORFs) constitute a new group of eukaryotic genes and are expected to represent 5% of the Drosophila melanogaster transcribed genes. In this review we provide a historical perspective of their recent discovery, describe their general mechanism and discuss the importance of smORFs for future genomic and transcriptomic studies. Finally, we discuss the biological role of the most studied smORF so far, the Mlpt/Pri/Tal gene in arthropods. The pleiotropic action of Mlpt/Pri/Tal in D. melanogaster suggests a complex evolutionary scenario that can be used to understand the origins, evolution and integration of smORFs into complex gene regulatory networks.
Tribolium; mlpt; pri; tarsal-less; Drosophila
Historical Perspective on the Discovery of small Open Reading Frames (smORFs)
Our knowledge of genome sequence, size and gene content has increased with the availability of new DNA sequencing technologies. This huge amount of data has opened new avenues for the development of bioinformatics. Bioinformatic prediction methods have been used to estimate the gene numbers of several eukaryotes, which vary considerably across groups. Estimations of gene contents do not support the theory that an increase in complexity can be associated with an increase in gene number. For example, a sponge genome contains more putative genes than a human genome does (Srivastava et al., 2010Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720–726.). However, transcriptome data have shown that half of the transcripts in mammalian genes are classified as non-coding RNAs (ncRNAs) because they do not contain large Open Reading Frames (ORFs) (e.g., Ota et al., 2004Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K et al. (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 36:40–45.).
In general, gene prediction methods attempt to identify intrinsic features in DNA sequences that are characteristics of exons, such as ORFs and Kozak consensus sequences (Figure 1). Traditionally, in silico approaches have considered a lower limit of 300 nucleotides or 100 amino acids for an ORF to be annotated as a putative exon. This in silico approach excludes small ORFs (smORFs) with fewer than 100 amino acids that might be biologically active. One of the first hints that smORFs might display a biological function was obtained by Kessler et al. (2003)Kessler MM, Zeng Q, Hogan S, Cook R, Morales AJ and Cottarel G (2003) Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res 13:264–271.. These authors identified new genes in S. cerevisiae by simply BLAST-searching potential budding yeast ORF products against sequences from other fungal and non-fungal species. Based on the hypothesis that conserved genes are functional, they found strong evidence for close to 100 new smORF genes in the S. cerevisiae genome (Kessler et al., 2003Kessler MM, Zeng Q, Hogan S, Cook R, Morales AJ and Cottarel G (2003) Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res 13:264–271.). Later, using functional genomics techniques, such as EST sequencing and mutant analysis, Kastenmayer et al. (2006)Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis,RW, Boeke JD et al. (2006) Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res 16:365–373. provided evidence for the existence of 299 smORFs; this figure represents approximately 5% of the annotated ORFs in S. cerevisiae. Furthermore, Kastenmayer et al. (2006)Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis,RW, Boeke JD et al. (2006) Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res 16:365–373. showed by specific gene deletion that 21 smORFs (∼8%) are essential for S. cerevisae viability. These smORFs are implicated in key cellular processes such as transport, intermediate metabolism and genome stability. Most importantly, at least some of the smORFs can be expressed and translated as peptides (Kastenmayer et al., 2006Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis,RW, Boeke JD et al. (2006) Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res 16:365–373.). Because several smORFs were shown to be conserved among fungi and higher eukaryotes (Kastenmayer et al., 2006Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis,RW, Boeke JD et al. (2006) Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res 16:365–373.), it remains to be investigated whether smORFs play a role during metazoan development.
Scheme of the general method for the identification of smORFs in different related species. Based on primary data and schemes from Kessler et al. (2003)Kessler MM, Zeng Q, Hogan S, Cook R, Morales AJ and Cottarel G (2003) Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res 13:264–271. and Ladoukakis et al. (2011)Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A and Couso JP (2011) Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol 12:R118.. smORF prediction is based on detection and filtering. The filtering process is important to reduce the false positive rate and increase the efficacy of functional smORFs estimation.
smORFs During Metazoan Development
Hormones and neuropeptides are considered the best examples of bioactive molecules of low molecular weight. They are transcribed as large mRNAs that are then processed into small peptides via post-translational mechanisms and proteolysis because they contain signal sequences at their N-termini. After processing in the ER and Golgi apparatus, hormones or neuropeptides can signal far from their production site (Figure 2A). Recent functional genomic studies have shown a new way of generating such small bioactive peptides, including the direct translation of smORFs. More than one smORF may be present in a single transcript. Hence, eukaryotic mRNA can be polycistronic, with multiple exons containing an initiation codon within a single mRNA (Savard et al., 2006Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569., Kondo et al., 2007Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665., Figure 2B). After secretion, smORFs act like hormones and neuropeptides and can be defined as a novel class of small peptide genes expressed during plant and animal morphogenesis (reviewed by Hashimoto et al., 2008Hashimoto Y, Kondo T and Kageyama Y (2008) Lilliputians get into the limelight: Novel class of small peptide genes in morphogenesis. Dev Growth Differ 50(Suppl 1):S269–276.).
Schematic drawings of the generation of biologically active short peptides. A similar scheme was published by Hashimoto et al. (2008)Hashimoto Y, Kondo T and Kageyama Y (2008) Lilliputians get into the limelight: Novel class of small peptide genes in morphogenesis. Dev Growth Differ 50(Suppl 1):S269–276.. (A) Hormones and neuropeptides are generated via a large mRNA precursor (blue) in the nucleus, then translated by ribosomes (green) from a single initiation codon and finally processed in the ER and Golgi into small peptides, which are subsequently secreted by vesicles to act far from the production site. (B) Polycistronic smORFs (red) can be translated by several ribosomes (green) along a single mRNA, followed by cell secretion. Peptides from smORFs can also act far from the releasing cell.
As previously mentioned, several smORFs have been identified based on their conserved structure and gene expression in fungi (Kessler et al., 2003Kessler MM, Zeng Q, Hogan S, Cook R, Morales AJ and Cottarel G (2003) Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res 13:264–271.; Kastenmayer et al., 2006Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis,RW, Boeke JD et al. (2006) Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res 16:365–373.). In plants, several smORFs were identified by using genetic screening methods such as POLARIS, ROTUN-DIFOLIA4, and Enod40. These plant smORFs encode peptides that are involved in morphogenetic processes, including root formation, leaf shape control, and cortical cell division during nodule formation (reviewed by Hashimoto et al., 2008Hashimoto Y, Kondo T and Kageyama Y (2008) Lilliputians get into the limelight: Novel class of small peptide genes in morphogenesis. Dev Growth Differ 50(Suppl 1):S269–276.). Because these smORFs have been found by unbiased genetic screenings and occupy small regions of plant genomes, it is likely that smORFs play a role in other biological processes and systems.
mille-pattes, tarsal-less and polished rice: The Same Gene Can Act in Different Developmental Contexts
smORFs may also play a major role in animal development. The first hint that a smORF was required for animal development was provided by Savard et al. (2006)Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569., who investigated the embryogenesis of the red flour beetle, Tribolium castaneum. In an EST screening, Savard et al. (2006)Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569. identified mille-pattes (mlpt), a polycistronic peptide encoding four smORFs, three of which containing an LDPTGLY domain. In Tribolium, mlpt acts like a bona fide gap gene because it regulates Hox genes (Savard et al., 2006Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569.). The down regulation of mlpt results in beetle larvae that have up to ten pairs of legs instead of the three pairs of legs observed in wild-type beetles. Moreover, mlpt is expressed in the thorax and at the posterior growth-zone, which is the region responsible for posterior segmentation in short-germ insects, including the beetle T. castaneum (Figure 3).
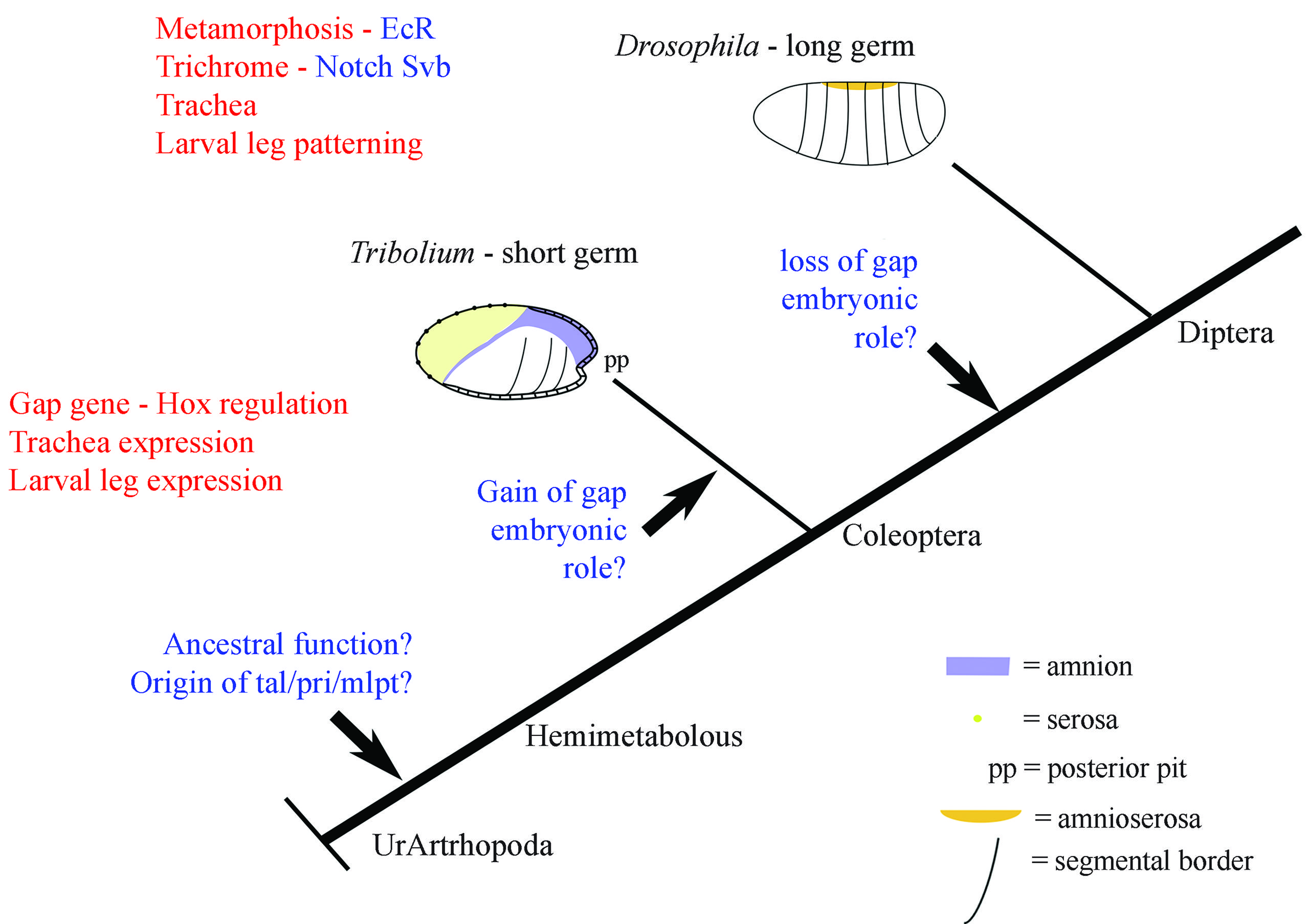
Evolution and functional role of Mlpt/Tal/Pri in arthropods. Several arthropods display an ortholog of Mlpt/Tal/Pri (original alignments and phylogenetic trees from Galindo et al., 2007Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106. and Savard et al., 2006Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569.). In the short-germ embryo of the beetle Tribolium castaneum, mlpt was shown to be expressed in the legs and trachea, where it acts as a gap gene during embryogenesis (Savard et al., 2006Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569.). In the long-germ embryo of the fly Drosophila melanogaster, Mlpt/Tal/Pri was shown to be involved in several processes, which are displayed in red (Chanut-Delalande et al., 2014Chanut-Delalande H, Hashimoto Y, Pelissier-Monier A, Spokony R, Dib A, Kondo T, Bohere J, Niimi K, Latapie Y, Inagaki S et al. (2014) Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol 16:1035–1044.; Galindo et al., 2007Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106.; Kondo et al., 2007Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665., 2010Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F and Kageyama Y (2010) Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329:336–339.; Pueyo and Couso, 2008Pueyo JI and Couso JP (2008) The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development. Dev Biol 324:192–201., 2011Pueyo JI and Couso JP (2011) Tarsal-less peptides control Notch signalling through the Shavenbaby transcription factor. Dev Biol 355:183–193.). Notch, Svb and EcR are the known regulators of Mlpt/Tal/Pri (Chanut-Delalande et al., 2014Chanut-Delalande H, Hashimoto Y, Pelissier-Monier A, Spokony R, Dib A, Kondo T, Bohere J, Niimi K, Latapie Y, Inagaki S et al. (2014) Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol 16:1035–1044.). Three unknown aspects of the evolution of Mlpt/Tal/Pri are highlighted in blue. These include the origin of the gene in arthropods, its ancestral function, and the loss of gap gene function after the split between the common ancestor of Coleoptera and Diptera. It is also possible that the gap gene function of Mlpt/Tal/Pri was independently acquired in Coleoptera.
Galindo et al. (2007)Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106. and Kondo et al. (2007)Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665. investigated the role of the mlpt ortholog in the fruit fly Drosophila melanogaster and provided interesting results regarding the function of these smORFs. Previously, this smORF ortholog was classified as a non-coding RNA in Drosophila because the small size of its ORFs suggested they were not translated (Tupy et al., 2005Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE and Rubin GM (2005) Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci USA 102:5495–5500..) The embryonic gap gene role of mlpt turned out not to be conserved in fruit flies, and synonyms of the same gene, such as tarsal-less (tal) (Galindo et al., 2007Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106.) or polished rice (pri) (Kondo et al., 2007Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665.) were found expressed in a pair-rule fashion during embryogenesis, but do not regulate Hox genes in flies. Kondo et al. (2007)Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665. also showed that pri is required non-cell autonomously and is essential for the formation of the specific F-actin bundles that will form the denticles, which are the typical epidermal structures of Drosophila larvae. In addition, pri is reported to function during tracheal morphogenesis (Kondo et al., 2007Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665.). Interestingly, the first four small peptides, which are similar to LDPTGLY, could be translated in an in vitro assay using S2 cells, but the last and largest peptide was not translated in this system (Kondo et al., 2007Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665.). In vivo rescue experiments have shown that these LDPTGLY peptides are functionally redundant and that the overexpression of one of these smORFs is able to rescue the denticle and the tracheal loss-of-function phenotype (Kondo et al., 2007Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665.). In addition to the roles reported during embryogenesis by Kondo et al. (2007)Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665., Galindo et al. (2007)Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106. provided evidence that tal is involved in leg patterning by demonstrating that tal is expressed at the leg imaginal discs and that tal hypomorphic mutants lack the whole tarsal region (Galindo et al., 2007Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106.).
These three initial studies opened new avenues into smORF research because they showed that a single smORF can be involved in several developmental contexts with apparently different biological roles during morphogenesis.
tal/pri/mlpt As a Case Study for the Biological Mechanism of a smORF
Though different biological roles for mlpt/tal/pri have been described in several developmental contexts, the mechanism by which these smORFs act during development remained unknown until quite recently. One of the first hints of the mechanism of action for mlpt/tal/pri was obtained by Kondo et al. (2010)Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F and Kageyama Y (2010) Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329:336–339., who investigated the specification and differentiation of larval epidermal denticle structures in Drosophila. It was previously shown that denticle differentiation in fruit flies is controlled by the activity of the transcription factor Shavenbaby (Svb) and its downstream target genes (Mevel-Ninio et al., 1995Mevel-Ninio M, Terracol R, Salles C, Vincent A and Payre F (1995) ovo, a Drosophila gene required for ovarian development, is specifically expressed in the germline and shares most of its coding sequences with shavenbaby, a gene involved in embryo patterning. Mech Dev 49:83–95.), Kondo et al. (2007)Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665. had shown that the loss of mlpt/tal/pri does not affect the expression of Svb, suggesting that these genes belong to different pathways that activate denticle formation. Subsequently, Kondo et al. (2010)Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F and Kageyama Y (2010) Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329:336–339. showed that the short 11 amino acid peptide found in mlpt/tal/pri is able to trigger the terminal truncation of Svb. This truncation converts Svb from a repressor that has accumulated in nuclear foci into a nucleoplasmic activator, both in vivo during denticle formation and in vitro in Drosophila S2 cells. This result was groundbreaking because it established that smORFs may cross the cell membrane and, upon reaching the nucleus, alter the function of essential transcription factors, such as Svb. This finding showed an important and new role for mlpt/tal/pri and, by association, for smORFs.
The biological functions of the mlpt/tal/pri genes have also been investigated in fruit fly leg specification, where they display two independent functions. The first function is in the determination of the presumptive tarsal region in early third instar larvae (Galindo et al., 2007Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106.; Pueyo and Couso, 2008Pueyo JI and Couso JP (2008) The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development. Dev Biol 324:192–201.). For tarsal determination, mlpt/tal/pri non-autonomously generates a new territory of presumptive tarsal cells by defining the presence of the transcription factors Rotund (Rn) and Spineless (Ss) and the absence of Dachshund (Dac) and B (Pueyo and Couso, 2008Pueyo JI and Couso JP (2008) The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development. Dev Biol 324:192–201.). Importantly, this role of tal-related peptides is independent of Svb, suggesting that mlpt/tal/pri peptides interact with partners other than Svb.
The second biological function of mlpt/tal/pri genes in the leg occurs later in development. During early pupal Drosophila development, Notch (N) signaling activates tal mRNA expression in stripes of cells in the distal part of each tarsal segment. Interestingly, the Tal peptides feed back on N signaling by repressing the transcription of Delta (Dl) in the tarsal joints. This feedback acts through the post-transcriptional activation of Svb in a similar manner to that described for trichomes during late embryogenesis (Mevel-Ninio et al., 1995Mevel-Ninio M, Terracol R, Salles C, Vincent A and Payre F (1995) ovo, a Drosophila gene required for ovarian development, is specifically expressed in the germline and shares most of its coding sequences with shavenbaby, a gene involved in embryo patterning. Mech Dev 49:83–95.; Delon et al., 2003Delon I, Chanut-Delalande H and Payre F (2003) The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech Dev 120:747–758.; Sucena et al., 2003Sucena E, Delon I, Jones I, Payre F and Stern DL (2003) Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424:935–938.). Thus, a common biological mechanism involving Notch signaling and Svb may control mlpt/pri/tal expression in several developmental contexts.
Finally, recent pioneering work has implicated the Mlpt/Pri/Tal peptides as mediators of ecdysone control of development (Chanut-Delalande et al., 2014Chanut-Delalande H, Hashimoto Y, Pelissier-Monier A, Spokony R, Dib A, Kondo T, Bohere J, Niimi K, Latapie Y, Inagaki S et al. (2014) Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol 16:1035–1044.). A previously uncharacterized enzyme of ecdysone biosynthesis in D. melanogaster, Glutathione S transferase E14 (GstE14) was shown to be required for mlpt/pri/tal expression. Moreover, the nuclear ecdysone receptor (EcR) was found to directly bind to the mlpt/pri/tal cis-regulatory region, which suggests a direct link between ecdysone action and mlpt/pri/tal activation (Chanut-Delalande et al., 2014Chanut-Delalande H, Hashimoto Y, Pelissier-Monier A, Spokony R, Dib A, Kondo T, Bohere J, Niimi K, Latapie Y, Inagaki S et al. (2014) Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol 16:1035–1044.). Therefore, Mlpt/Pri/Tal peptides provide a molecular framework to explain how systemic hormonal signaling is able to execute different genetic programs both throughout embryonic development and post-embryonically (Chanut-Delalande et al., 2014Chanut-Delalande H, Hashimoto Y, Pelissier-Monier A, Spokony R, Dib A, Kondo T, Bohere J, Niimi K, Latapie Y, Inagaki S et al. (2014) Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol 16:1035–1044.).
Based on these essential roles of mlpt/pri/tal in several contexts, it is important to estimate how many other smORFs may play a role in other developmental processes. This has only recently been addressed using the new bioinformatic and molecular biology techniques described below (Ladoukakis et al., 2011Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A and Couso JP (2011) Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol 12:R118.; Aspden et al., 2014Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M and Couso JP (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3:e03528.).
How Many smORFs Exist in Animal Genomes? Lessons From Fruit Flies
Quantifying how many smORFs exist within animal genomes is not trivial because the prediction methods used to identify coding sequence are biased against detecting very short open reading frames (< 100 bp) (e.g., Saeys et al., 2007Saeys Y, Rouze P and Van de Peer Y (2007) In search of the small ones: Improved prediction of short exons in vertebrates, plants, fungi and protists. Bioinformatics 23:414–420.). In general, gene prediction methods use either a de novo approach with mathematical models that determine the probabilities for all possible intron-exon annotations in a given sequence, or a comparison to a known genome or cDNA sequences from related organisms (Ladoukakis et al., 2011Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A and Couso JP (2011) Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol 12:R118.). smORFs that contain fewer than 100 amino acids and correspond to functional genes may not be predicted and can thus be grouped with non-functional smORFs that can occur by chance (Windsor and Mitchell-Olds, 2006Windsor AJ and Mitchell-Olds T (2006) Comparative genomics as a tool for gene discovery. Curr Opin Biotechnol 17:161–167.). Ladoukakis et al. (2011)Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A and Couso JP (2011) Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol 12:R118. used a comparative approach to investigate the smORFs of the fruit fly species D. melanogaster and D. pseudoobscura, two related species that are separated from their common ancestor by 25 to 55 million years. This investigation led to a range of between 401 and 4.561 functional smORFs in Drosophila. In fact, 401 smORFs would represent 3% of the 13,907 protein-coding genes that have been annotated as of 2011 (FlyBase release 5; as accessed in October 2011). Thus, a substantial number of biologically relevant smORFs await characterization.
A detailed functional analysis of one of these candidates, the transcript encoded by the gene putative non-coding RNA003in2L (pncr003:2L), indicated that this gene contains two potentially functional smORFs of 28 and 29 amino acids in a single sequence, which led to exciting results (Magny et al., 2013Magny G, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA and Couso JP (2013) Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341:1116–1120.). pcnr003:2L regulates calcium transport and thus influences regular muscle contraction in the Drosophila heart (Magny et al., 2013Magny G, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA and Couso JP (2013) Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341:1116–1120.). In contrast to the mlpt/pri/tal peptides, which are small and do not display a clear secondary structure, such as a alpha-helix or beta-sheet, pncr003:2L peptides have a predicted helical structure. Searches for a structural homolog have identified two paralogs in the human genome, sarcolamban (Scl) and phospholambam (Pcl), which both contain two smORFs of 30 amino acids that are similar to pncr003:2L. Functional analysis of these human homologs indicates that they play a conserved role in calcium trafficking, particularly in regulating the activity of the sarco-endoplasmic reticulum Ca2+ adenosine triphosphatase (SERCA) enzyme. Thus, these smORF peptides are required for regular muscle contraction in humans and Drosophila (Magny et al., 2013Magny G, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA and Couso JP (2013) Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341:1116–1120.).
Future Directions and Open Questions in smORF Research
Although considerable progress has been made in smORF research over the past few years, as highlighted by this review, several questions remain open. First, it is not known how many smORFs are important for developmental processes. Sequence analysis has shown that hundreds of smORFs are conserved among Drosophila species, suggesting that a large number of smORFs are functional (Ladoukakis et al., 2011Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A and Couso JP (2011) Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol 12:R118.). As 401 of these conserved smORFs are also expressed during Drosophila embryogenesis, it is likely that these smORFs are functional, because high expression levels are suggestive of a functional role. It is expected that several other smORFs will have their function analyzed, in addition to mlpt/pri/tal (Galindo et al., 2007Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106.; Kondo et al., 2007Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665.; Savard et al., 2006Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569.) and pncr003:2L (Magny et al., 2013Magny G, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA and Couso JP (2013) Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341:1116–1120.).
A new promising technique, Poly-Ribo-Seq, was recently applied in the experimental validation and discovery of new smORFs (Aspden et al., 2014Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M and Couso JP (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3:e03528.). Briefly, Poly-Ribo-Seq requires polysome isolation for the determination of the sequence bound by each of the ribosomes. Polysomes are clusters of multiple ribosomes that are bound to mRNA during translation. The Poly-Ribo-Seq approach thus reduces the number of false positives and doubled the number of annotated smORFs in Drosophila S2 cells (Aspden et al., 2014Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M and Couso JP (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3:e03528.), thereby increasing the evidence of translation from 107 to 228 smORFs. By using this approach, 700 functional smORFs were estimated within the Drosophila genome by Aspden et al. (2014)Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M and Couso JP (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3:e03528..
Recently, Lu et al., 2014Lu Y, Zhuang Y and Liu J (2014) Mining antimicrobial peptides from small open reading frames in Ciona intestinalis. J Peptide Sci 20:25–29. synthesized ten bioactive peptides from the smORFs found in the genome of the ascidian Ciona intestinalis and tested them as potential antimicrobial peptides (AMPs). Five of these peptides were active against bacterial strains, suggesting that they may act as antimicrobial peptides (AMPs) in ascidians. Thus, it is possible that clusters of smORFs are activated upon infection in a fast response and release.
What other open questions exist in smORF research? One of the most important regards the evolution of the most described smORF, mlpt/pri/tal in arthropods (Savard et al., 2006Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569.; Galindo et al., 2007Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106.; Kondo et al., 2007Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665.). Because mlpt/pri/tal is involved in several biological processes, such as early patterning, trichome, tracheal patterning and leg patterning, and was recently shown to be involved in metamorphosis (Chanut-Delalande et al., 2014Chanut-Delalande H, Hashimoto Y, Pelissier-Monier A, Spokony R, Dib A, Kondo T, Bohere J, Niimi K, Latapie Y, Inagaki S et al. (2014) Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol 16:1035–1044.), it will be important to investigate the evolutionary origin and ancestral role of this smORF (Figure 3). Is mlpt/pri/tal involved in all of these biological processes also in hemimetabolous insects and other arthropods? Evolutionary studies on mlpt/pri/tal have the potential to contribute to the discussion about the interaction between genetic developmental control and the environment, the so-called Eco-Evo-Devo field of knowledge (Abouheif et al., 2014Abouheif E, Fave MJ, Ibarraran-Viniegra AS, Lesoway MP, Rafiqi AM and Rajakumar R (2014) Eco-evo-devo: The time has come. Adv Exp Med Biology 781:107–125.). If 5% of the genes in a given genome are smORFs, as recently suggested for Drosophila melanogaster (Aspden et al., 2014Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M and Couso JP (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3:e03528.), it will be interesting to investigate whether at least some other developmental pathways such as Hh, Wnt, FGFs and BMPs are also regulated by and regulate other smORFs.
Importantly, it will be interesting to know whether smORFs are found in basal metazoans such as sponges, cnidarians and ctenophores, as the examples described so far are primarily restricted to yeast, plants, arthropods and chordates. Additionally, we expect that, as experimental and bioinformatic methods become more powerful, smORFs will be essential components of genome annotations and studies of gene regulatory networks. Finally, examples of horizontal smORF transfer between eubacteria and eukaryotes and parasites might be discovered.
Acknowledgments
The RNdF lab is supported by Fundação de Amparo de Pesquisa do Estado do Rio de Janeiro (FAPERJ), Conselho Nacional de Pesquisa (CNPq) and Fundação Educacional de Macaé (FUNEMAC). JPA is a master student of Programa de Pós-Graduação em Produtos Bioativos e Biociências (PPG-PRODBIO), VTS is supported by a PIBIC/CNPq scholarship and ACR by a FAPERJ scholarship.
-
Associate Editor: Igor Schneider
References
- Abouheif E, Fave MJ, Ibarraran-Viniegra AS, Lesoway MP, Rafiqi AM and Rajakumar R (2014) Eco-evo-devo: The time has come. Adv Exp Med Biology 781:107–125.
- Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M and Couso JP (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3:e03528.
- Chanut-Delalande H, Hashimoto Y, Pelissier-Monier A, Spokony R, Dib A, Kondo T, Bohere J, Niimi K, Latapie Y, Inagaki S et al. (2014) Pri peptides are mediators of ecdysone for the temporal control of development. Nat Cell Biol 16:1035–1044.
- Delon I, Chanut-Delalande H and Payre F (2003) The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila Mech Dev 120:747–758.
- Galindo MI, Pueyo JI, Fouix S, Bishop SA and Couso JP (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106.
- Hashimoto Y, Kondo T and Kageyama Y (2008) Lilliputians get into the limelight: Novel class of small peptide genes in morphogenesis. Dev Growth Differ 50(Suppl 1):S269–276.
- Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis,RW, Boeke JD et al. (2006) Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae Genome Res 16:365–373.
- Kessler MM, Zeng Q, Hogan S, Cook R, Morales AJ and Cottarel G (2003) Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res 13:264–271.
- Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665.
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F and Kageyama Y (2010) Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329:336–339.
- Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A and Couso JP (2011) Hundreds of putatively functional small open reading frames in Drosophila Genome Biol 12:R118.
- Lu Y, Zhuang Y and Liu J (2014) Mining antimicrobial peptides from small open reading frames in Ciona intestinalis J Peptide Sci 20:25–29.
- Magny G, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA and Couso JP (2013) Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341:1116–1120.
- Mevel-Ninio M, Terracol R, Salles C, Vincent A and Payre F (1995) ovo, a Drosophila gene required for ovarian development, is specifically expressed in the germline and shares most of its coding sequences with shavenbaby, a gene involved in embryo patterning. Mech Dev 49:83–95.
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K et al. (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 36:40–45.
- Pueyo JI and Couso JP (2008) The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development. Dev Biol 324:192–201.
- Pueyo JI and Couso JP (2011) Tarsal-less peptides control Notch signalling through the Shavenbaby transcription factor. Dev Biol 355:183–193.
- Saeys Y, Rouze P and Van de Peer Y (2007) In search of the small ones: Improved prediction of short exons in vertebrates, plants, fungi and protists. Bioinformatics 23:414–420.
- Savard J, Marques-Souza H, Aranda M and Tautz D (2006) A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569.
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720–726.
- Sucena E, Delon I, Jones I, Payre F and Stern DL (2003) Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424:935–938.
- Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE and Rubin GM (2005) Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster Proc Natl Acad Sci USA 102:5495–5500.
- Windsor AJ and Mitchell-Olds T (2006) Comparative genomics as a tool for gene discovery. Curr Opin Biotechnol 17:161–167.
Publication Dates
-
Publication in this collection
Sept 2015
History
-
Received
14 Jan 2015 -
Accepted
30 Mar 2015