Abstract
The distribution of 45S rDNA cluster in avian karyotypes varies in different aspects, such as position, number of bearer chromosomes, and bearers being macro- or microchromosomes. The present study investigated the patterns of variation in the 45S rDNA-bearer chromosomes of birds in order to understand the evolutionary dynamics of the cluster configuration and its contribution to the evolution of bird karyotypes. A total of 73 bird species were analyzed, including both published data and species for which rDNA-FISH was conducted for the first time. In most birds, the 45S rDNA clusters were located in a single pair of microchromosomes. Hence, the location of 45S rDNA in macrochromosomes, observed only in Neognathae species, seems to be a derived state, probably the result of chromosomal fusion between microchromosomes and distinct macrochromosomes. Additionally, the 45S rDNA was observed in multiple microchromosomes in different branches of the bird phylogeny, suggesting recurrence of dispersion processeses, such as duplications and translocations. Overall, this study indicated that the redistribution of the 45S rDNA sites in bird chromosomes followed different evolutionary trajectories with respect to each lineage of the class Aves.
Keywords:
FISH; chromosome; chromosome evolution; cytogenetics; Aves
Introduction
The rDNA genes are extremely important for cell function, given that they encode the rRNA involved in ribosome biogenesis (Hadjiolov, 1985Hadjiolov AA (1985) The Nucleolus and Ribosome Biogenesis. Springer Verlag, Wien.; Shaw and Brown, 2012Shaw P and Brown J (2012). Nucleoli: Composition, function, and dynamics. Plant Physiol 158:44-51.). In this process, two rDNA clusters are involved: the 45S rDNA composed by 18S, 5.8S, and 28S genes, and internal (ITS1 and ITS2) and external (5’ETS and 3’ETS) transcribed spacers; and the 5S rDNA, composed by a 5S gene separated by an intergenic spacer region (IGS) (Daniels and Delany, 2003Daniels LM and Delany ME (2003) Molecular and cytogenetic organization of the 5S ribosomal DNA array in chicken (Gallus gallus). Chromosome Res 11:305-317.; Dyomin et al., 2016Dyomin AG, Koshel EI, Kiselev AM, Saifitdinova AF, Galkina SA, Fukagawa T, Kostareva AA and Gagiskaya ER (2016) Chicken rRNA gene cluster structure. PLoS One 11:e0157464.). In the eukaryotic genome, multiple copies of these clusters are organized in tandem in the DNA, forming the 5S and 45S rDNA sites in the chromosome (Daniels and Delany, 2003Daniels LM and Delany ME (2003) Molecular and cytogenetic organization of the 5S ribosomal DNA array in chicken (Gallus gallus). Chromosome Res 11:305-317.; Dyomin et al., 2016Dyomin AG, Koshel EI, Kiselev AM, Saifitdinova AF, Galkina SA, Fukagawa T, Kostareva AA and Gagiskaya ER (2016) Chicken rRNA gene cluster structure. PLoS One 11:e0157464.).
Identification of chromosomes that bear 45S rDNA can be performed by the silver nitrate impregnation technique (Ag-NOR) (Howell and Black, 1980Howell WM and Black DA (1980) Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia 36:1014-1015.). However, this procedure only identifies the chromosomes with 45S rDNA sites in transitional activity, exhibiting intercellular, and interindividual variation (Zurita et al., 1997Zurita F, Sánchez A, Burgos M, Jiménez R and de la Guardia RD (1997) Interchromosomal, intercellular and interindividual variability of NORs studied with silver staining and in situ hybridization. Heredity 78:229-234.). In this way, fluorescence in situ hybridization (FISH) experiments are more appropriate for this type of study, since they allow the precise identification of the bearing chromosomes when using probes for the genes that make up the rDNA cluster even when they are not active (O’Connor, 2008O’Connor C (2008) Fluorescence in situ hybridization (FISH). Nat Educ 1:171.).
In recent years, FISH has been increasingly used to detect rDNA-bearer chromosomes in a range of vertebrate and invertebrate species (e.g., Roy et al., 2005Roy V, Monti-Dedieu L, Chaminade N, Siljak-Yakovlev S, Aulard S, Lemeunier F and Montchamp-Moreau C (2005) Evolution of the chromosomal location of rDNA genes in two Drosophila species subgroups: ananassae and melanogaster. Heredity 94:388–395.; Cazaux et al., 2011Cazaux B, Catalan J, Veyrunes F, Douzery EJ and Britton-Davidian J (2011) Are ribosomal DNA clusters rearrangement hotspots? a case study in the genus Mus (Rodentia, Muridae). BMC Evol Biol 11:124.; Mazzoleni et al., 2018Mazzoleni S, Rovatsos M, Schillaci O and Dumas F (2018) Evolutionary insight on localization of 18S, 28S rDNA genes on homologous chromosomes in Primates genomes. Comp Cytogenet 12:27-40.; Sochorová et al., 2018Sochorová J, Garcia S, Gálvez F, Symonová R and Kovarík A (2018) Evolutionary trends in animal ribosomal DNA loci. introduction to a new online database. Chromosoma 127:141-150.). These studies have shown that 45S and 5S rDNA sites are most frequently found in a single chromosome pair per diploid genome, although considerable variation has been observed, with up to 74 chromosome copies for the 5S rDNA cluster sites and 54 for the 45S (Sochorová et al., 2018Sochorová J, Garcia S, Gálvez F, Symonová R and Kovarík A (2018) Evolutionary trends in animal ribosomal DNA loci. introduction to a new online database. Chromosoma 127:141-150.). In addition, no significant correlation has been found between the number of 5S and 45S loci, which suggests that their distribution and amplification within the karyotype follow independent evolutionary trajectories (Sochorová et al., 2018Sochorová J, Garcia S, Gálvez F, Symonová R and Kovarík A (2018) Evolutionary trends in animal ribosomal DNA loci. introduction to a new online database. Chromosoma 127:141-150.).
The location of rDNA sites has been related to hotspots of chromosomal breakage (Cazaux et al., 2011Cazaux B, Catalan J, Veyrunes F, Douzery EJ and Britton-Davidian J (2011) Are ribosomal DNA clusters rearrangement hotspots? a case study in the genus Mus (Rodentia, Muridae). BMC Evol Biol 11:124.). This fragility is probably originated by the repetitive nature of clusters or their intense gene expression activity (Huang et al., 2008Huang J, Ma L, Yang F, Fei SZ and Li L (2008) 45S rDNA regions are chromosome fragile sites expressed as gaps in vitro on metaphase chromosomes of root-tip meristematic cells in Lolium spp. PLoS One 3:e2167.). In the chromosome, these breakages may result in different types of rearrangements, such as translocation, fusions, duplications, and inversions, leading to rapid changes in the chromosomal distribution of the rDNA sites in closely related species (Datson and Murray, 2006Datson PM and Murray BG (2006) Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome Res 14:845-857.; Degrandi et al., 2014Degrandi TM, Pita S, Panzera Y, de Oliveira EHC, Marques JRF, Figueiró MR, Marques LC, VInadé L, Gunski RJ and Del Valle Garnero A (2014) Karyotypic evolution of ribosomal sites in buffalo subspecies and their crossbreed. Genet Mol Biol 37:375-380).
Birds are a highly diversified biological group with more than 10,000 species. On the other hand, less than 12% of the species have a known karyotype (Kretschmer et al., 2018aKretschmer R, Ferguson-Smith MA and de Oliveira EHC (2018a) Karyotype evolution in birds: From conventional staining to chromosome painting. Genes 9:E181.). The diploid number ranges from 2 n= 40, as found in Burhinus oedicnemus, to 2n = 136-142 in Corythaixoides concolor (Christidis, 1990Christidis L (1990) Animal cytogenetics 4: Chordata 3 B: Aves. Gebrüder Borntraeger, Berlin, pp 55–57.; Nie et al., 2009Nie W, O’Brien PCM, Fu BL, Beiyuan NG, Vitaly V, Carter NP, Ferguson-Smith MA and Yang F (2009) Avian comparative genomics: reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes) - an atypical species with low diploid number. Chromosome Res 17:99-113.). However, the karyotype of birds is relatively conserved, and most species have 2n = 80. Generally, their karyotypes are characterized by the presence of macrochromosomes, which are 2.5–6 μm in length, and microchromosomes, which are less than 2.5 μm long (Rodionov, 1996Rodionov AV (1996) Micro vs. macro: A review of structure and functions of avian micro- and macrochromosomes. Russ J Genet 32:517–527.; Kretschmer et al., 2018aKretschmer R, Ferguson-Smith MA and de Oliveira EHC (2018a) Karyotype evolution in birds: From conventional staining to chromosome painting. Genes 9:E181.). This basic karyotype structure can be seen in the species of both the Paleognathae and Neognathae clades (Kretschmer et al., 2018aKretschmer R, Ferguson-Smith MA and de Oliveira EHC (2018a) Karyotype evolution in birds: From conventional staining to chromosome painting. Genes 9:E181.).
Studies that have mapped the chromosomal location of 45S rDNA sites have shown considerable divergence among birds (Nishida-Umehara et al., 2007Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y and Griffin K (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in Palaeognathous birds. Chromosome Res 15:721-734.; Nishida et al., 2008Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, Griffin DK and Matsuda Y (2008) Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res 16:171-181., 2013Nishida C, Ishijima J, Ishishita S, Yamada K, Griffin DK, Yamazaki T and Matsuda Y (2013) Karyotype reorganization with conserved genomic compartmentalization in dot-shaped microchromosomes in the Japanese mountain Hawk-eagle (Nisaetus nipalensis orientalis, Accipitridae). Cytogenet Genome Res 141:284-294.; Nie et al., 2009Nie W, O’Brien PCM, Fu BL, Beiyuan NG, Vitaly V, Carter NP, Ferguson-Smith MA and Yang F (2009) Avian comparative genomics: reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes) - an atypical species with low diploid number. Chromosome Res 17:99-113.; Tagliarini et al., 2009Tagliarini MM, Pieczarka JC, Nagamachi CY, Rissino J and de Oliveira EHC (2009) Chromosomal analysis in Cathartidae: Distribution of heterochromatic blocks and rDNA, and phylogenetic considerations. Genetica 135:299-304.; de Oliveira et al., 2013de Oliveira EHC, Tagliarini MM, dos Santos MS, O’ Brien PCM and Ferguson-Smith MA (2013) Chromosome painting in three species of Buteoninae: A cytogenetic signature reinforces the monophyly of south American species. PLoS One 8: e70071.; Kretschmer et al., 2014Kretschmer R, Gunski RJ, Garnero ADV, Furo IDO, O’Brien PCM, Ferguson-Smith MA and de Oliveira EH (2014) Molecular cytogenetic characterization of multiple intrachromosomal rearrangements in two representatives of the genus Turdus (Turdidae, Passeriformes). PLoS One 9:e103338.; Degrandi et al., 2017Degrandi TM, Garnero ADV, O’Brien PCM, Ferguson-Smith MA, Kretschmer R, de Oliveira EHC and Gunski RJ (2017) Chromosome painting in Trogon s. surrucura (Aves, Trogoniformes) reveals a karyotype derived by chromosomal fissions, fusions, and inversions. Cytogenet Genome Res 151:208-215.; de Oliveira et al., 2017de Oliveira TD, Kretschmer R, Bertocchi NA, Degrandi TM, de Oliveira EHC, Cioffi MDB, Garnero AD and Gunski RJ (2017) Genomic organization of repetitive DNA in woodpeckers (Aves, Piciformes): implications for karyotype and ZW sex chromosome differentiation. PLoS One 12:e169987.). In Paleognathae birds, the 45S rDNA is normally found in a single microchromosome pair (Nishida-Umehara et al., 2007Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y and Griffin K (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in Palaeognathous birds. Chromosome Res 15:721-734.). However, in the Neognathae birds, a significant variation has been observed, including species with 45S rDNA clusters in multiple microchromosomes, in a single macrochromosome pair, or in both (Nishida et al., 2008Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, Griffin DK and Matsuda Y (2008) Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res 16:171-181.; de Oliveira et al., 2013de Oliveira EHC, Tagliarini MM, dos Santos MS, O’ Brien PCM and Ferguson-Smith MA (2013) Chromosome painting in three species of Buteoninae: A cytogenetic signature reinforces the monophyly of south American species. PLoS One 8: e70071.; Tagliarini, 2013Tagliarini MM (2013) Aplicação de pintura cromossômica em espécies da família Accipitridae (Aves, Falconiformes): Considerações filogenéticas e evolutivas. D. Sc. Thesis, Universidade Federal do Pará, Belém, http://repositorio.ufpa.br/jspui/handle/2011/5839.
http://repositorio.ufpa.br/jspui/handle/...
; Degrandi et al., 2017Degrandi TM, Garnero ADV, O’Brien PCM, Ferguson-Smith MA, Kretschmer R, de Oliveira EHC and Gunski RJ (2017) Chromosome painting in Trogon s. surrucura (Aves, Trogoniformes) reveals a karyotype derived by chromosomal fissions, fusions, and inversions. Cytogenet Genome Res 151:208-215.; de Oliveira et al., 2017de Oliveira TD, Kretschmer R, Bertocchi NA, Degrandi TM, de Oliveira EHC, Cioffi MDB, Garnero AD and Gunski RJ (2017) Genomic organization of repetitive DNA in woodpeckers (Aves, Piciformes): implications for karyotype and ZW sex chromosome differentiation. PLoS One 12:e169987.). However, the origin of this variation and its possible evolutionary implications are still poorly understood.
Thus, the aim of this study was to investigate this variation in 45S rDNA-bearing chromosomes of birds in order to understand the evolutionary dynamics of the cluster configuration and its contribution to the evolution of the bird karyotype.
Materials and Methods
Specimens
In this work, we analyzed the basic karyotype structure and distribution of the 45S rDNA sites in bird karyotypes. The following data were considered in each species: diploid number, number of 45S rDNA-bearing chromosomes, their type (macro- or microchromosome), and position of the clusters on the chromosome arm. First, the data were obtained from the literature, considering only the species in which the 45S rDNA clusters were identified by FISH-rDNA. Ag-NORs data were disregarded due to the intercellular and individual variations or possible false positive results, already reported in the literature.
Additionally, 29 species were selected from the sample bank of the Laboratory of Animal Genetic Diversity at Universidade Federal do Pampa for the first rDNA-FISH screening of each taxon: order Passeriformes/family Thraupidae: Tachyphonus coronatus, Coryphospingus cucullatus; Icteridae: Agelaioides badius, Molothrus bonariensis, Tyrannidae: Pitangus sulphuratus, Myiarchus ferox; Tityridae: Schiffornis virescens; Furnariidae: Dendrocolaptes platyrostris, Anumbius annumbi, Synallaxis albescens, Furnarius rufus, Cranioleuca obsoleta, Syndactila rufosuperciliata; Coraciiformes/ Alcedinidae: Chloroceryle americana; Piciformes/Ramphastidae: Ramphastos tucanus; Accipitriformes/Accipitridae: Pseudastur albicollis, Buteogallus urubitinga; Pelecaniformes/Ardeidae: Syrigma sibilatrix; Charadriiformes/Stercorariidae: Stercorarius antarcticus; Caprimulgiformes /Trochilidae: Amazilia versicolor, Nyctibiidae: Nyctibius griseus, Caprimulgidae: Hydropsalis torquata; Cuculiformes/Cuculidae: Coccyzus melacoryphus, Piaya cayana, Guira guira; Columbiformes/Columbidae: Columbina talpacoti; Tinamiformes/ Tinamidae: Nothura maculosa and Rhynchotus rufescens (Table 1).
Chromosome preparation
Mitotic chromosomes were obtained following standard protocols, including direct preparation from bone marrow, fibroblast culture, and lymphocyte culture (Moorhead et al., 1960Moorhead OS, Nowell PC, Mellinan WJ, Battips DM and Ungerford DA (1960) Chromossome preparations of leukocytes cultured from human peripheral blood. Exp Cell Res 20:613-616.; Sasaki et al., 1968Sasaki M, Ikeuchi T and Maino S (1968) A feather pulp culture for avian chromosomes with notes on the chromosomes of the peafowl and the ostrich. Experientia 24:1923-1929.; Garnero and Gunski, 2000Garnero ADV and Gunski RJ (2000) Comparative analysis of the karyotypes of Nothura maculosa and Rynchotus rufescens (Aves: Tinamidae). A case of chromosomal polymorphism. Nucleus 43:64-70.).
FISH for 18S rDNA
FISH using probes specific for the 18S rDNA gene identified the 45S rDNA-bearing chromosomes. Primers were developed from sequences obtained from the fish Hoplias malabaricus (Cioffi et al., 2009Cioffi MB, Martins C, Centofante L, Jacobina U and Bertollo LAC (2009) Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: Mapping of three classes of repetitive DNAs. Cytogenet Genome Res 125:132-141.). This generated a fragment of approximately 1,400 base pairs, which was labeled by polymerase chain reaction (PCR), using the primers 18SF (5’CCGAGGACCTCACTAAACCA 3’) and 18SR (5’CCGCTTTGGTGACTCTTGAT-3’), with fluorescein dUTP in the PCR mix.
The PCRs were run in a final volume of 25 μL containing 2 ng of genomic DNA from H. malabaricus, 0.2 μM of each primer (18SF and 18SR), 0.2 mM of dNTP, 10X buffer (1x), 50 mM of MgCl2 (2 μM), 1 mM of Fluorescein-12-dUTP solution, 1 U/μL of Taq polymerase, and sterile H2O to complete to final volume. The thermal cycling parameters were 94 ºC for 60 s, 30 cycles of 94 ºC for 60 s, 60 ºC for 60 s, 72 ºC for 90 s, followed by an elongation step of 5 min at 72 ºC (Cioffi et al., 2009Cioffi MB, Martins C, Centofante L, Jacobina U and Bertollo LAC (2009) Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: Mapping of three classes of repetitive DNAs. Cytogenet Genome Res 125:132-141.).
For the FISH procedures, slides with metaphases were treated with RNase A (10 μg/mL) for 20 min and then denatured in 70% formamide at 70°C for 80 s. Subsequently, 300 ng of the 18S probe were added to each slide, which was then sealed with a cover slip and incubated overnight at 37 °C (Daniels and Delany, 2003Daniels LM and Delany ME (2003) Molecular and cytogenetic organization of the 5S ribosomal DNA array in chicken (Gallus gallus). Chromosome Res 11:305-317.). The slides were then washed in 50% formamide at 42 °C for 1 min (x2), 2xSSC at 40 °C for 2.5 min (x2), and once in 4xSSC Tween (1X) at room temperature. The chromosomes were counterstained with DAPI. Hybridization results were analyzed using a Zeiss Axioplan2 fluorescence microscope.
Chromosomal analyses
The diploid number of each specimen was determined from the analysis of approximately 30 mitotic cells stained with Giemsa observed under an optical microscope. Variation in the number of rDNA clusters was evaluated based on the number of chromosomes that presented a fluorescent signal. The rDNA cluster-bearing chromosomes were classified as either macrochromosomes or microchromosomes, according to their length (Rodionov, 1996Rodionov AV (1996) Micro vs. macro: A review of structure and functions of avian micro- and macrochromosomes. Russ J Genet 32:517–527.). The position of the 45S rDNA cluster was classified as: (i) pericentromeric (adjacent to the centromere), (ii) subtelomeric (adjacent to the telomere), and (iii) interstitial (between the centromere and the telomere) (Cazaux et al., 2011Cazaux B, Catalan J, Veyrunes F, Douzery EJ and Britton-Davidian J (2011) Are ribosomal DNA clusters rearrangement hotspots? a case study in the genus Mus (Rodentia, Muridae). BMC Evol Biol 11:124.). Ideograms were created using these characteristics to represent the rDNA-bearing chromosomes in each species.
Phylogenetic comparison
The species were compared using the phylogenetic relationships proposed by Jarvis et al. (2014)Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT et al. (2014) Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320-1331. and Prum et al. (2015)Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM and Lemmon AR (2015) A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526:569-573.. In this step, the chromosomal locations of the 45S rDNA clusters were plotted in a modified phylogenetic tree of Jarvis et al. (2014)Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT et al. (2014) Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320-1331.. In this tree, we used the Mesquite software to exclude groups of birds for which rDNA location data were not available. We considered the presence of 45S rDNA in a single pair of microchromosomes as an ancestral condition for birds, according to the hypothesis of Nishida-Umehara et al. (2007)Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y and Griffin K (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in Palaeognathous birds. Chromosome Res 15:721-734.. Based on this hypothesis, we analyzed the evolutionary relationships and the probable chromosomal rearrangements that would explain the variations observed in chromosomes carrying 45S rDNA.
Results
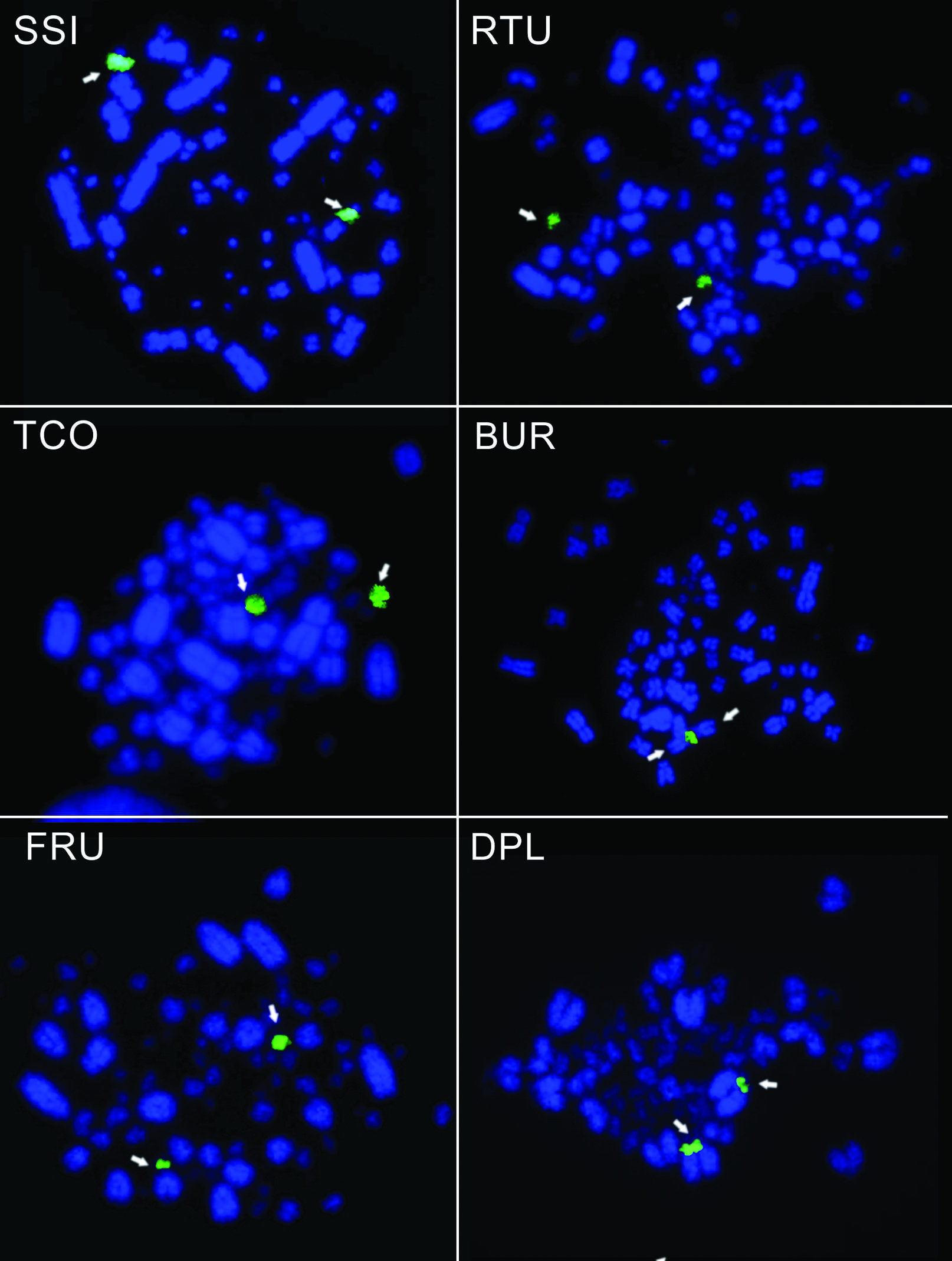
The number of chromosomes (2n), number of 45S rDNA sites, and the characteristics of these bearing chromosomes from 29 selected species for rDNA-FISH screening in this work are shown in Table 1 (see species identified as ‘present study’ in Table 1). The rDNA-FISH results of some selected species are shown in Figure 1.
Examples of the metaphases analyzed in the present study using the 18S rDNA probe (green) to identify the chromosomes (blue) carrying 45S rDNA sites (arrows). The acronym shown in the upper right corner of each metaphase indicates the species: Syrigma sibilatrix (SSI), Ramphastos tucanus (RTU), Tachyphonus coronatus (TCO), Buteogallus urubitinga (BUR), Furnarius rufus (FRU), and Dendrocolaptes platyrostris (DPL).
Overall, the analysis of the chromosomal distribution of the 45S rDNA included 73 bird species, representing 17 orders of the class Aves (Table 1). Eight of these species were Paleognaths, representing four orders, the Casuariiformes, Rheiformes, Struthioniformes, and Tinamiformes. The other 65 species were Neognaths, belonging to 13 orders, the Accipitriformes, Caprimulgiformes, Charadriiformes, Columbiformes, Coraciiformes, Cuculiformes, Falconiformes, Galliformes, Passeriformes, Pelecaniformes, Piciformes, Psittaciformes, and Trogoniformes.
Variation in the diploid number in birds
Considering only the bird species for which the location of 45S rDNA sites is available (73), diploid numbers ranged from 2n = 40 to 2n = 112 (Table 1). Despite this ample variation, most (38) of the species had diploid numbers between 78 and 82, and 21 were 2n = 80 (Figure 2A). While the Paleognathae species were relatively conserved, with most species having around 80 chromosomes, higher variability in 2n was observed in Neognathae (Table 1).
Chromosomal location of the 45S rDNA sites in all 73 bird species analyzed in the present study. (A) variation in the diploid number; (B) variation in the number of 45S rDNA bearer chromosomes; (C) the proportion of the species with 45S rDNA located in macrochromosomes or microchromosome; (D) location of the 45S rDNA cluster in the chromosome arm.
Number of 45S rDNA sites
The analysis of the number of 45S rDNA-bearing chromosomes highlighted that most (58) species had a cluster in a single chromosome pair (Figure 2B). In the Paleognathae, Nothura maculosa and Eudromia elegans were exceptions, with two rDNA-bearing chromosome pairs. In the Neognathae, the 45S rDNA clusters were found in a single chromosome pair and in up to six or seven pairs (Table 1).
Types of rDNA-bearing chromosomes
In the bimodal analysis of macrochromosomes vs. microchromosomes, the 45S rDNA sites of most (59) species were observed on microchromosomes (Figure 2C). In the Paleognathae, the rDNA was located exclusively on microchromosomes. The Neognathae presented different configurations, by contrast, with some species having the cluster in the microchromosomes, others in the macrochromosomes, and some in both types of chromosome, as observed in the Accipitriformes, Harpia harpija (Table 1).
The location of the rDNA in macrochromosomes was observed in 14 Neognathae species (Table 1), representing a number of different orders: Pandion haliaetus, Pseudastur albicollis, Buteogallus urubitinga, Buteo nitidus, Rupornis magnirostris, B. meridionalis, H. harpyja and Morphnus guianensis (Accipitriformes), Burhinus oedicnemus (Charadriiformes), Piaya cayana and Guira guira (Cuculiformes), Dendrocolaptes platyrostris (Passeriformes), Colaptes campestres, and Colaptes melanochloros (Piciformes). In some cases, it was possible to identify homologies between the macrochromosomes and those of Gallus gallus (Table 2).
Associations of 45S rDNA sites with macrochromosomes and their respective homologies with Gallus gallus (GGA) chromosomes.
Position of the 45S rDNA site in the chromosomes
As microchromosomes have a limited resolution, the species with rDNA sites in these tiny elements were excluded from the analysis of the rDNA topology in the chromosomes in order to avoid biases in data interpretation. Therefore, the position of the rDNA cluster was analyzed only in the 14 species in which the 45S rDNA is located in macrochromosomes.
The 45S rDNA was observed in a pericentromeric position in most (64%) cases, that is, in P. haliaetus, P. albicollis, B. urubitinga, B. nitidus, R. magnirostris, B. meridionalis (Accipitriformes), G. guira, P. cayana (Cuculiformes), and D. platirostris (Passeriformes). The interstitial position was the second most frequent, being observed in 22% of the species, B. oedicnemus (Charadriiformes), C. campestres, and C. melanochloros (Piciformes). Finally, a subtelomeric position was recorded in two (14%) species, M. guianensis and H. harpyja (Accipitriformes) (Figure 2D, Table 1).
Phylogenetic comparisons
For the phylogenetic comparisons, the presence of the 45S rDNA cluster in a single pair of microchromosomes was considered to be the ancestral condition, based on the hypothesis of Nishida-Umehara et al. (2007)Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y and Griffin K (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in Palaeognathous birds. Chromosome Res 15:721-734.. This analysis revealed that the variation in the number of 45S rDNA-bearer chromosomes was independent of the phylogenetic relationships among the species (Figure 3). The presence of rDNA in macrochromosomes was observed in species belonging to different orders from infra class Neognathae (Figure 3).
Phylogenetic relationships among the birds modified from Jarvis et al., 2014Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT et al. (2014) Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320-1331.. The data of chromosomal location of the 45S rDNA from species analyzed in the present study were plotted in the tree. (A) Species with rDNA located only in a microchromosome pair; (B) species with rDNA in multiples microchromosomes; (C) species in which the rDNA is located in macrochromosomes. The complete data are shown in Table 1.
Discussion
Here we present for the first time a broad analysis of the distribution of 45S rDNA in avian karyotype. Although an impressive variation was observed in the chromosomes carrying the 45S rDNA cluster, we recorded that in most species it is located in a single pair of microchromosomes. Interestingly, most of these species have a karyotype with 2n = 80 chromosomes (Figure 2A).
A study with rodents indicated that there is no relationship between the 2n and the number of 45S rDNA cluster bearing chromosomes (Cazaux et al., 2011Cazaux B, Catalan J, Veyrunes F, Douzery EJ and Britton-Davidian J (2011) Are ribosomal DNA clusters rearrangement hotspots? a case study in the genus Mus (Rodentia, Muridae). BMC Evol Biol 11:124.). Nevertheless, birds with 2n = 80 chromosomes that carry only a single pair of 45S rDNA microchromosomes seem to reflect the karyotype conservation status of these species in relation to the ancestral karyotype of birds (PAK), as proposed by Griffin et al. (2007)Griffin DK, Robertson LB, Tempest HG and Skinner BM (2007) The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res 117: 64-77.. This karyotype uniformity of birds has also been observed in species from Paleognathae and Neognathae using the GGA whole chromosome paint (Kretschmer et al., 2018aKretschmer R, Ferguson-Smith MA and de Oliveira EHC (2018a) Karyotype evolution in birds: From conventional staining to chromosome painting. Genes 9:E181.).
The presence of a single pair of microchromosomes with 45S rDNA conserved among the species of Paleognaths (Dromaius novaehollandiae, Casuarius casuarius, Struthio camelus, Rhea pennata, and Rhea americana) suggests that this would be an ancestral condition of rDNA (Nishida-Umehara et al., 2007Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y and Griffin K (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in Palaeognathous birds. Chromosome Res 15:721-734.). Using the phylogenetic relationships proposed by Jarvis et al. (2014)Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT et al. (2014) Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320-1331. and Prum et al. (2015)Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM and Lemmon AR (2015) A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526:569-573., we compared these data and identified that several species of the Neognaths infraclass preserve the 45S rDNA in a pair of microchromosome (Figure 3), a fact that reinforces the hypothesis of PAK ancestral condition (Griffin et al., 2007Griffin DK, Robertson LB, Tempest HG and Skinner BM (2007) The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res 117: 64-77.).
The 45S rDNA-bearer chromosome is related to the presence or absence of the process of karyotypic diversification. For example, Accipitriformes, where species of the Cathartidae family have karyotypes with 80 chromosomes, the 45S rDNA was located in only a single pair of microchromosomes (Raudsepp et al., 2002Raudsepp T, Houck ML, O’Brien PMC, Ferguson-Smith MA, Ryder OA and Chowdhary BP (2002) Cytogenetic analysis of California Condor (Gymnogyps californianus) chromosomes: comparison with chicken (Gallus gallus) macrochromosomes. Cytogenet Genome Res 98:54-60.; Tagliarini et al., 2009Tagliarini MM, Pieczarka JC, Nagamachi CY, Rissino J and de Oliveira EHC (2009) Chromosomal analysis in Cathartidae: Distribution of heterochromatic blocks and rDNA, and phylogenetic considerations. Genetica 135:299-304.). In contrast, the Accipitridae family shows a diploid number quite derived (2n = 58-82), and chromosome painting evidenced an extensive karyotypic reorganization, originated by breaks and fusions of macrochromosomes (GGA) and microchromosomes. In this group, it was observed that 45S rDNA is associated with different macrochromosomes (Table 2) (de Oliveira et al., 2013de Oliveira EHC, Tagliarini MM, dos Santos MS, O’ Brien PCM and Ferguson-Smith MA (2013) Chromosome painting in three species of Buteoninae: A cytogenetic signature reinforces the monophyly of south American species. PLoS One 8: e70071., Tagliarini, 2013Tagliarini MM (2013) Aplicação de pintura cromossômica em espécies da família Accipitridae (Aves, Falconiformes): Considerações filogenéticas e evolutivas. D. Sc. Thesis, Universidade Federal do Pará, Belém, http://repositorio.ufpa.br/jspui/handle/2011/5839.
http://repositorio.ufpa.br/jspui/handle/...
; Nishida et al., 2014Nishida C, Ishishita S, Yamada K, Griffin DK and Matsuda Y (2014) Dynamic chromosome reorganization in the Osprey (Pandion haliaetus, Pandionidae, Falconiformes): Relationship between chromosome size and the chromosomal distribution of centromeric repetitive DNA sequences. Cytogenet Genome Res 142:179-189.).
45S rDNA in multiple microchromosomes
Multiple microchromosomes carrying 45S rDNA can be found in some species of the orders Tinamiformes, Columbiformes, Trogoniformes, and Falconiformes, and notably, even phylogenetically related species may differ in the number of rDNA bearing chromosomes. For instance, Paleognath birds from the order Tinamiformes show variation in the number of clusters. In R. rufescens a single microchromosome pair containing the 45S rDNA was observed, whereas in N. maculosa and E. elegans, the 45S rDNA is located in two pairs of microchromosomes (Figure 3). Similarly, such numerical variation is also seen in species of the same genus, as in the genus Falco (Falconiformes), where F. tinnunculus has 45S rDNA in four microchromosome pairs, F. columbarius in five pairs, and F. peregrinus shows this cluster in six or seven pairs (Nishida et al., 2008Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, Griffin DK and Matsuda Y (2008) Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res 16:171-181.) (Figure 3). Considering the phylogenetic relationships between these orders, the most plausible explanation for the origin of these variation are recurrent processes of 45S rDNA cluster duplications or translocations, resulting in the numerical variation observed in these species.
45S rDNA distribution in macrochromosomes
The 45S rDNA location in macrochromosomes can be considered a derived characteristic in birds (Kretschmer et al., 2018aKretschmer R, Ferguson-Smith MA and de Oliveira EHC (2018a) Karyotype evolution in birds: From conventional staining to chromosome painting. Genes 9:E181.). The available data on chromosomal homologies with G. gallus (GGA) (Table 2), demonstrated that the rDNA sites are clearly associated with distinct macrochromosomes. This scenario might have been originated by multiple independent events of chromosomal fusion, which are supported by several different types of evidence.
In Accipitriformes, for example, multiple associations were recorded, including GGA1, GGA3, and GGA7. In B. nitidus, R. magnirostris, and B. meridionallis, an association with the homologous GGA7 segment was found, although the short arm of the chromosome pair containing the rDNA of these species was not hybridized by any of the GGA probes used (de Oliveira et al., 2013de Oliveira EHC, Tagliarini MM, dos Santos MS, O’ Brien PCM and Ferguson-Smith MA (2013) Chromosome painting in three species of Buteoninae: A cytogenetic signature reinforces the monophyly of south American species. PLoS One 8: e70071.). This unhybridized region probably corresponds to the homologous of the ancestral microchromosome containing the rDNA, reinforcing the fusion hypothesis. Similarly, in P. haliaetus, the rDNA located on the q-arm of chromosome 2 was associated with the homologous GGA1 segment (Nishida et al., 2014Nishida C, Ishishita S, Yamada K, Griffin DK and Matsuda Y (2014) Dynamic chromosome reorganization in the Osprey (Pandion haliaetus, Pandionidae, Falconiformes): Relationship between chromosome size and the chromosomal distribution of centromeric repetitive DNA sequences. Cytogenet Genome Res 142:179-189.). In this species, the short arm did not hybridize by any GGA probe. However, P. haliaetus showed rDNA in the long arm, suggesting that a pericentric inversion should have occurred after fusion with the 45S rDNA microchromosome, shifting the cluster position to the long arm.
45S rDNA related to intrachromosomal rearrangements
Intrachromosomal rearrangements have been reported in bird karyotypes, and our data revealed that two cases involved the 45S rDNA-bearer chromosome (Degrandi et al., 2017Degrandi TM, Garnero ADV, O’Brien PCM, Ferguson-Smith MA, Kretschmer R, de Oliveira EHC and Gunski RJ (2017) Chromosome painting in Trogon s. surrucura (Aves, Trogoniformes) reveals a karyotype derived by chromosomal fissions, fusions, and inversions. Cytogenet Genome Res 151:208-215.). For example, in Cuculiformes, Piaya caiana and Guira guira showed the association of 45S rDNA with a segment homologue to chromosome GGA2 (Table 2). In P. caiana, the cluster was in the pericentromeric region of the short arm of the submetacentric chromosome pair 7, whereas in G. guira the cluster was in the long arm pericentromeric region of the metacentric chromosome 6 (Figure 3). In Accipitriformes Harpia harpyja and Pandion haliaetus, the association was with a segment homologue to chromosome GGA1 (Table 2). However, in H. harpyja, the rDNA cluster was seen in the subtelomeric region of macrochromosome 6, and in P. haliaetus, the cluster occupied the pericentromeric region of the long arm on chromosome 2 (Figure 3) (Tagliarini, 2013Tagliarini MM (2013) Aplicação de pintura cromossômica em espécies da família Accipitridae (Aves, Falconiformes): Considerações filogenéticas e evolutivas. D. Sc. Thesis, Universidade Federal do Pará, Belém, http://repositorio.ufpa.br/jspui/handle/2011/5839.
http://repositorio.ufpa.br/jspui/handle/...
; Nishida et al., 2014Nishida C, Ishishita S, Yamada K, Griffin DK and Matsuda Y (2014) Dynamic chromosome reorganization in the Osprey (Pandion haliaetus, Pandionidae, Falconiformes): Relationship between chromosome size and the chromosomal distribution of centromeric repetitive DNA sequences. Cytogenet Genome Res 142:179-189.). The translocation or a pericentric inversion may explain this position variation of the internal 45S rDNA cluster in the bearer chromosome, which corroborates the hypothesis that the 45S rDNA cluster is related to chromosomal breakpoints, according to Cazaux et al. (2011)Cazaux B, Catalan J, Veyrunes F, Douzery EJ and Britton-Davidian J (2011) Are ribosomal DNA clusters rearrangement hotspots? a case study in the genus Mus (Rodentia, Muridae). BMC Evol Biol 11:124..
Conclusion
In birds, the 45S rDNA site is located predominantly in a single pair of microchromosomes, although a number of deviations from this basic pattern exist, with some species having rDNA located in more than one microchromosome pair or in macrochromosomes, or in both types of chromosome. The present study also demonstrated that the redistribution of rDNA sites within the chromosome complement has resulted from chromosomal rearrangements, which have resulted from the distinct evolutionary histories of each group of the class Aves.
Acknowledgments
We would like to thank all our colleagues from the Grupo de Pesquisa Diversidade Genética Animal from Universidade Federal do Pampa (UNIPAMPA), Laboratório Cultura de Tecidos e Citogenética (SAMAM) from Instituto Evandro Chagas, and the Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior - Brasil (CAPES) - Finance Code 001.
References
- Cazaux B, Catalan J, Veyrunes F, Douzery EJ and Britton-Davidian J (2011) Are ribosomal DNA clusters rearrangement hotspots? a case study in the genus Mus (Rodentia, Muridae). BMC Evol Biol 11:124.
- Christidis L (1990) Animal cytogenetics 4: Chordata 3 B: Aves. Gebrüder Borntraeger, Berlin, pp 55–57.
- Cioffi MB, Martins C, Centofante L, Jacobina U and Bertollo LAC (2009) Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: Mapping of three classes of repetitive DNAs. Cytogenet Genome Res 125:132-141.
- Daniels LM and Delany ME (2003) Molecular and cytogenetic organization of the 5S ribosomal DNA array in chicken (Gallus gallus). Chromosome Res 11:305-317.
- Datson PM and Murray BG (2006) Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome Res 14:845-857.
- Degrandi TM, Pita S, Panzera Y, de Oliveira EHC, Marques JRF, Figueiró MR, Marques LC, VInadé L, Gunski RJ and Del Valle Garnero A (2014) Karyotypic evolution of ribosomal sites in buffalo subspecies and their crossbreed. Genet Mol Biol 37:375-380
- Degrandi TM, Garnero ADV, O’Brien PCM, Ferguson-Smith MA, Kretschmer R, de Oliveira EHC and Gunski RJ (2017) Chromosome painting in Trogon s. surrucura (Aves, Trogoniformes) reveals a karyotype derived by chromosomal fissions, fusions, and inversions. Cytogenet Genome Res 151:208-215.
- de Oliveira EHC, Tagliarini MM, Rissino JD, Pieczarka JC, Nagamachi CY, O’Brien PCM and Ferguson-Smith MA (2010) Reciprocal chromosome painting between white hawk (Leucopternis albicollis) and chicken reveals extensive fusions and fissions during karyotype evolution of Accipitridae (Aves, Falconiformes). Chromosome Res 18:349-355.
- de Oliveira EHC, Tagliarini MM, dos Santos MS, O’ Brien PCM and Ferguson-Smith MA (2013) Chromosome painting in three species of Buteoninae: A cytogenetic signature reinforces the monophyly of south American species. PLoS One 8: e70071.
- de Oliveira TD, Kretschmer R, Bertocchi NA, Degrandi TM, de Oliveira EHC, Cioffi MDB, Garnero AD and Gunski RJ (2017) Genomic organization of repetitive DNA in woodpeckers (Aves, Piciformes): implications for karyotype and ZW sex chromosome differentiation. PLoS One 12:e169987.
- dos Santos MS, Kretschmer R, Silva FA, Ledesma MA, O’ Brien PCM, Ferguson-Smith MA, Del Valle Garnero A, de Oliveira EH and Gunski RJ (2015) Intrachromosomal rearrangements in two representatives of the genus Saltator (Thraupidae, Passeriformes) and the occurrence of heteromorphic Z chromosomes. Genetica 143:535-543.
- dos Santos MDS, Kretschmer R, Vilches CF, Bakker A, Gahr M, O’Brien PCM, Ferguson Smith MA and de Oliveira EHC (2017) Comparative cytogenetics between two important songbird, models: the zebra finch and the canary. PLoS One 12:e170997.
- Dyomin AG, Koshel EI, Kiselev AM, Saifitdinova AF, Galkina SA, Fukagawa T, Kostareva AA and Gagiskaya ER (2016) Chicken rRNA gene cluster structure. PLoS One 11:e0157464.
- Garnero ADV and Gunski RJ (2000) Comparative analysis of the karyotypes of Nothura maculosa and Rynchotus rufescens (Aves: Tinamidae). A case of chromosomal polymorphism. Nucleus 43:64-70.
- Griffin DK, Robertson LB, Tempest HG and Skinner BM (2007) The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res 117: 64-77.
- Hadjiolov AA (1985) The Nucleolus and Ribosome Biogenesis. Springer Verlag, Wien.
- Howell WM and Black DA (1980) Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia 36:1014-1015.
- Huang J, Ma L, Yang F, Fei SZ and Li L (2008) 45S rDNA regions are chromosome fragile sites expressed as gaps in vitro on metaphase chromosomes of root-tip meristematic cells in Lolium spp PLoS One 3:e2167.
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT et al. (2014) Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320-1331.
- Kretschmer R, Gunski RJ, Garnero ADV, Furo IDO, O’Brien PCM, Ferguson-Smith MA and de Oliveira EH (2014) Molecular cytogenetic characterization of multiple intrachromosomal rearrangements in two representatives of the genus Turdus (Turdidae, Passeriformes). PLoS One 9:e103338.
- Kretschmer R, de Oliveira EHC, dos Santos MS, Furo IDO, O’Brien PCM, Ferguson-Smith MA, del Valle Garnero and Gunski RJ (2015) Chromosome mapping of the large Elaenia (Elaenia spectabilis): evidence for a cytogenetic signature for passeriform birds? Biol J Linn Soc 115:391-398.
- Kretschmer R, Ferguson-Smith MA and de Oliveira EHC (2018a) Karyotype evolution in birds: From conventional staining to chromosome painting. Genes 9:E181.
- Kretschmer R, de Oliveira TD, Furo IO, Silva FAO, Gunski RJ, Garnero AV, Cioffo MB, de Oliveira EHC and de Freitas TRO (2018b) Repetitive DNAs and shrink genomes: a chromosomal analysis in nine Columbidae species (Aves, Columbiformes). Genet Mol Biol 41:98-106.
- Mazzoleni S, Rovatsos M, Schillaci O and Dumas F (2018) Evolutionary insight on localization of 18S, 28S rDNA genes on homologous chromosomes in Primates genomes. Comp Cytogenet 12:27-40.
- McPherson MC, Robinson CM, Gehlen LP and Delany ME (2014) Comparative cytogenomics of Poultry: Mapping of single gene and repeat loci in the Japanese quail (Coturnix japonica). Chromosome Res 22:71-83.
- Moorhead OS, Nowell PC, Mellinan WJ, Battips DM and Ungerford DA (1960) Chromossome preparations of leukocytes cultured from human peripheral blood. Exp Cell Res 20:613-616.
- Nie W, O’Brien PCM, Fu BL, Beiyuan NG, Vitaly V, Carter NP, Ferguson-Smith MA and Yang F (2009) Avian comparative genomics: reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes) - an atypical species with low diploid number. Chromosome Res 17:99-113.
- Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, Griffin DK and Matsuda Y (2008) Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res 16:171-181.
- Nishida C, Ishijima J, Ishishita S, Yamada K, Griffin DK, Yamazaki T and Matsuda Y (2013) Karyotype reorganization with conserved genomic compartmentalization in dot-shaped microchromosomes in the Japanese mountain Hawk-eagle (Nisaetus nipalensis orientalis, Accipitridae). Cytogenet Genome Res 141:284-294.
- Nishida C, Ishishita S, Yamada K, Griffin DK and Matsuda Y (2014) Dynamic chromosome reorganization in the Osprey (Pandion haliaetus, Pandionidae, Falconiformes): Relationship between chromosome size and the chromosomal distribution of centromeric repetitive DNA sequences. Cytogenet Genome Res 142:179-189.
- Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y and Griffin K (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in Palaeognathous birds. Chromosome Res 15:721-734.
- O’Connor C (2008) Fluorescence in situ hybridization (FISH). Nat Educ 1:171.
- Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM and Lemmon AR (2015) A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526:569-573.
- Raudsepp T, Houck ML, O’Brien PMC, Ferguson-Smith MA, Ryder OA and Chowdhary BP (2002) Cytogenetic analysis of California Condor (Gymnogyps californianus) chromosomes: comparison with chicken (Gallus gallus) macrochromosomes. Cytogenet Genome Res 98:54-60.
- Rodionov AV (1996) Micro vs. macro: A review of structure and functions of avian micro- and macrochromosomes. Russ J Genet 32:517–527.
- Roy V, Monti-Dedieu L, Chaminade N, Siljak-Yakovlev S, Aulard S, Lemeunier F and Montchamp-Moreau C (2005) Evolution of the chromosomal location of rDNA genes in two Drosophila species subgroups: ananassae and melanogaster Heredity 94:388–395.
- Sasaki M, Ikeuchi T and Maino S (1968) A feather pulp culture for avian chromosomes with notes on the chromosomes of the peafowl and the ostrich. Experientia 24:1923-1929.
- Seibold-Torres C, Owens E, Chowdhary R, Ferguson-Smith MA, Tizard I and Raudsepp T (2015) Comparative cytogenetics of the Congo African Grey Parrot (Psittacus erithacus). Cytogenet Genome Res 147:144-153.
- Shaw P and Brown J (2012). Nucleoli: Composition, function, and dynamics. Plant Physiol 158:44-51.
- Sochorová J, Garcia S, Gálvez F, Symonová R and Kovarík A (2018) Evolutionary trends in animal ribosomal DNA loci. introduction to a new online database. Chromosoma 127:141-150.
- Tagliarini MM, Pieczarka JC, Nagamachi CY, Rissino J and de Oliveira EHC (2009) Chromosomal analysis in Cathartidae: Distribution of heterochromatic blocks and rDNA, and phylogenetic considerations. Genetica 135:299-304.
- Zurita F, Sánchez A, Burgos M, Jiménez R and de la Guardia RD (1997) Interchromosomal, intercellular and interindividual variability of NORs studied with silver staining and in situ hybridization. Heredity 78:229-234.
Internet Resources
- Gill F and D Donsker (2018) IOC World Bird List (v 8.1), https://www.worldbirdnames.org/ioc-lists/crossref/
» https://www.worldbirdnames.org/ioc-lists/crossref/ - Tagliarini MM (2013) Aplicação de pintura cromossômica em espécies da família Accipitridae (Aves, Falconiformes): Considerações filogenéticas e evolutivas. D. Sc. Thesis, Universidade Federal do Pará, Belém, http://repositorio.ufpa.br/jspui/handle/2011/5839
» http://repositorio.ufpa.br/jspui/handle/2011/5839 - Mesquite: a modular system for evolutionary analysis - Version 3.40, http://mesquiteproject.org
» http://mesquiteproject.org
-
Associate Editor: Yatiyo Yonenaga-Yassuda
Publication Dates
-
Publication in this collection
30 Mar 2020 -
Date of issue
2020
History
-
Received
28 Nov 2018 -
Accepted
08 May 2019