Abstract
Genetically antimicrobial resistance in Mycobacterium tuberculosis is currently one of the most important aspects of tuberculosis, considering that there are emerging resistant strains for almost every known drug used for its treatment. There are multiple antimicrobials used for tuberculosis treatment, and the most effective ones are the first-line drugs, which include isoniazid, pyrazinamide, rifampicin, and ethambutol. In this context, understanding the mechanisms of action and resistance of these molecules is essential for proposing new therapies and strategies of treatment. Additionally, understanding how and where mutations arise conferring a resistance profile to the bacteria and their effect on bacterial metabolism is an important requisite to be taken in producing safer and less susceptible drugs to the emergence of resistance. In this review, we summarize the most recent literature regarding novel mutations reported between 2017 and 2022 and the advances in the molecular mechanisms of action and resistance against first-line drugs used in tuberculosis treatment, highlighting recent findings in pyrazinamide resistance involving PanD and, additionally, resistance-conferring mutations for novel drugs such as bedaquiline, pretomanid, delamanid and linezolid.
Keywords:
Mycobacterium tuberculosis
; antimicrobial resistance; isoniazid; pyrazinamide; ethambutol; rifampicin
Introduction
Tuberculosis (TB) is an infectious disease caused predominantly by Mycobacterium tuberculosis (Mtb). This disease is a serious problem for public health since it afflicted about 10 million people worldwide, which culminated in 1.3 million deaths only in 2020 This makes TB the second most common cause of death by a single infectious agent, only surpassed in recent years by COVID-19. Among the most used medicines in the treatment of TB, isoniazid (INH), pyrazinamide (PZA), ethambutol (EMB), and rifampicin (RIF) are called first-line drugs (Figure 1). These drugs are the first choice of treatment for TB, which has a regimen of about six months with co-administration of all of them in the first four months and two of them in the last two ones (World Health Organization, 2021World Health Organization (2021) Global Tuberculosis Report 2021. ).
First-line drugs used in tuberculosis treatment. Structures for first-line drugs used in tuberculosis treatment, Isoniazid, Pyrazinamide, Rifampicin and Ethambutol. Structures were drawn using Marvin software (Cherinka et al., 2019Cherinka B, Andrews BH, Sánchez-Gallego J, Brownstein J, Argudo-Fernández M, Blanton M, Bundy K, Jones A, Masters K, Law DR et al. (2019) Marvin: A tool kit for streamlined access and visualization of the SDSS-IV MaNGA data set. Astron J 158:74.).
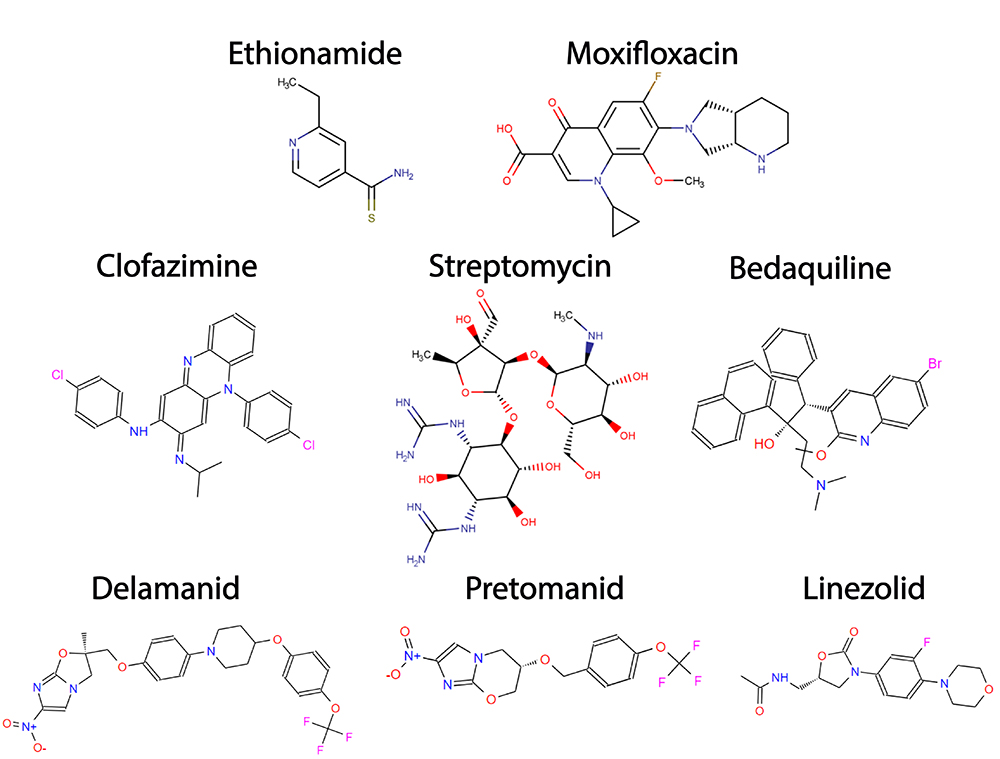
Additionally, to the first-line drugs, other antimicrobials, including ethionamide (ETH), injectable aminoglycosides, fluoroquinolones, diarylquinolines, and nitroimidazoles can also be used, but only against resistant strains. These antimicrobials are denominated as second-line drugs (Figure 2).The second-line drugs have been proven to have lower efficacy and higher toxicity compared to first-line drugs and require a longer regimen of treatment that could take more than a year (Wolff and Nguyen, 2012Wolff KA and Nguyen L (2012) Strategies for potentiation of ethionamide and folate antagonists against Mycobacterium tuberculosis. Expert Rev Anti Infect Ther 10:971-981.; Gopal and Dick, 2014Gopal P and Dick T (2014) Reactive dirty fragments: Implications for tuberculosis drug discovery. Curr Opin Microbiol 21:7-12.; World Health Organization, 2021World Health Organization (2021) Global Tuberculosis Report 2021. ). Nevertheless, new regimens, based on novel or repurposed drugs with anti-TB activity such as bedaquiline, delamanid, pretomanid, linezolid, clofazimine and moxifloxacin are currently in phase III clinical trials, aiming to reduce or simplify the current chemotherapy for MDR-TB and XDR-TB some of which are ZeNix, endTB and SimpliciTB. (Perrin et al., 2022Perrin C, Athersuch K, Elder G, Martin M and Alsalhani A (2022) Recently developed drugs for the treatment of drug-resistant tuberculosis: A research and development case study. BMJ Glob Heal 7:e007490.).
Second-line drugs used in tuberculosis treatment. Structures for some second-line drugs used in tuberculosis treatment, Ethionamide, Moxifloxacin (Fluoroquinolone), Clofazimine, Streptomycin (injectable aminoglycoside), Bedaquiline, Delamanid, Pretomanid and Linezolid Structures were drawn using Marvin software (Cherinka et al., 2019Li K, Yang Z, Gu J, Luo M, Deng J and Chen Y (2021) Characterization of pncA Mutations and Prediction of PZA Resistance in Mycobacterium tuberculosis Clinical Isolates From Chongqing, China. Front Microbiol 11:594171.).
Worrisomely, because of the long treatment, which includes severe side effects that contribute to the non-effective adhesion of the regimen, the number of genetically resistant and multiresistant strains to all in-use drugs against TB is alarmingly high and increases every year. The resistance to antimicrobials in TB is predominantly caused by either intrinsic resistance (particularly because of the complex mycobacterial cell wall and the presence of a chromosomal β-lactamase) or by mutations in genes (including promotor and encoding regions) from the antimicrobials targets and/or key enzymes for activating pro-drugs, such as INH and PZA. Plasmid horizontal transference is not reported so far in Mtb and consequently, this is not considered an important aspect of mycobacteria antimicrobial resistance (Smith et al., 2012Sinha P, Srivastava GN, Tripathi R, Mishra MN and Anupurba S (2020) Detection of mutations in the rpoB gene of rifampicin-resistant Mycobacterium tuberculosis strains inhibiting wild type probe hybridization in the MTBDR plus assay by DNA sequencing directly from clinical specimens. BMC Microbiol 20:284.; Zhang and Yew, 2015Zhang Y and Yew WW (2015) Mechanisms of drug resistance in Mycobacterium tuberculosis: Update 2015. Int J Tuberc Lung Dis 19:1276-1289. ). Mono resistance is common for INH, RIF and even streptomycin, however, many resistant strains of Mtb are resistant to at least two drugs. Based on that, the resistance in TB can be classified into multiresistant strains (MDR-TB), which consists of those strains that are resistant at least to INH and RIF; Pre-extensively drug-resistant TB (pre-XDR-TB), which includes those strains resistant to INH, RIF, a fluoroquinolone and a further injectable second-line drug, such as aminoglycosides; and extensive resistant strains (XDR-TB), which carry on all MDR-TB resistances and further resistances to at least one drug from the fluoroquinolone group combined with resistance to a group A drug, such as bedaquiline, levofloxacin, moxifloxacin or linezolid (World Health Organization, 2021World Health Organization (2021) Global Tuberculosis Report 2021. ).
In this review, we highlight the genetic mechanisms of resistance identified in Mtb for the first-line drugs, including INH, PZA, RIF and EMB. Additionally, although this is not the main focus of this revision, we also succinctly discuss the mechanisms of action and resistance involved against the most important second-line drugs, including bedaquiline, pretomanid, linezolid and clofazimine. We discuss the findings aiming to understand the mechanisms of resistance in this bacteria, as well as the recently reported polymorphisms and resistance-conferring mutations described in the last 5 years for first-line drugs used in the active TB treatment. This gathering of information has a pivotal role in proposing new strategies for more personalized treatment and improving clinical practices contributing to avoiding the dissemination of MDR, Pre-XDR, and XDR strains.
Resistance to INH
Isoniazid or isonicotinic acid hydrazide (INH) (Figure 1) is one of the most efficient anti-TB drugs (World Health Organization, 2021World Health Organization (2021) Global Tuberculosis Report 2021. ) and has been used as an anti-tubercular agent since 1952 (Selikoff et al., 1952Seifert M, Catanzaro D, Catanzaro A and Rodwell TC (2015) Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: A systematic review. PLoS One 10:e0119628.). INH is formed by a hydrazine group attached to a pyridine moiety and is considered a narrow-spectrum antimicrobial with bactericidal activity against Mtb during the actively-growing bacterial phase and bacteriostatic against slow-growing and latent stage (Fernandes et al., 2017Fernandes GF dos S, Salgado HRN and Santos JL dos (2017) Isoniazid: A review of characteristics, properties and analytical methods. Crit Rev Anal Chem 47:298-308.). INH is largely used against TB and it is one of the antimicrobials used in the standard TB regimen treatment (Sotgiu et al., 2015Somoskovi A, Parsons LM and Salfinger M (2001) The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir Res 2:164-168.; World Health Organization, 2021World Health Organization (2021) Global Tuberculosis Report 2021. ). INH was first identified as a groundbreaking anti-TB agent in 1951, and since then has been widely used against TB (Murray et al., 2015Musser JM, Kapur V, Williams DL, Kreiswirth BN, Van Soolingen D and Van Embden JDA (1996) Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: Restricted array of mutations associated with drug resistance. J Infect Dis 173:196-202.). Because of that, it is not a surprise that INH resistance is identified in more than 11% of all TB cases (World Health Organization, 2021World Health Organization (2021) Global Tuberculosis Report 2021. ).
INH is a pro-drug and needs to be activated by a catalase-peroxidase system, particularly by the enzyme KatG (Zhang et al., 1992Zhang Y, Heym B, Allen B, Young D and Cole S (1992) The catalase - Peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593.). KatG oxidizes INH in two steps: during the first one occurs the formation of isonicotinoyl radical; and in the second one, the radical reacts with ammonia to form isonicotinamide, the active state of INH (Bodiguel et al., 2001Bodiguel J, Nagy JM, Brown KA and Jamart-Grégoire B (2001) Oxidation of isoniazid by manganese and Mycobacterium tuberculosis catalase-peroxidase yields a new mechanism of activation. J Am Chem Soc 123:3832-3833.; Pierattelli et al., 2004Pierattelli R, Banci L, Eady NAJ, Bodiguel J, Jones JN, Moody PCE, Raven EL, Jamart-Grégoire B and Brown KA (2004) Enzyme-catalyzed mechanism of isoniazid activation in class I and class III peroxidases. J Biol Chem 279:39000-39009.; Timmins et al., 2004Timmins GS, Master S, Rusnak F and Deretic V (2004) Nitric oxide generated from isoniazid activation by KatG: Source of nitric oxide and activity against Mycobacterium tuberculosis. Antimicrob Agents Chemother 48:3006-3009.) (Figure 3). In addition to the active form of INH, several reactive oxygen species are also produced during this conversion (Shoeb et al., 1985Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, Wang H, Zhang W and Zhang Y (2011) Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis: A potential mechanism for shortening the duration of tuberculosis chemotherapy. Science 333:1630-1632.). When activated, INH forms an adduct with NADH, the INH-NAD, through the formation of a covalent bond with the nicotinamide group of this coenzyme (Figure 3). INH-NAD is the responsible molecule for inhibiting the biosynthesis of mycolic acids in Mtb by binding to 2-trans-enoyl-acyl carrier protein reductase (InhA). This enzyme has a Rossmanoid fold and belongs to the NADH-dependent short dehydrogenase/reductase family and catalyzes the reduction of trans-2-enoyl-ACP fatty acids (Dessen et al., 1998) (Figure 4D). As an enzyme from the fatty acid system II (FAS-II), InhA is particularly involved in the elongation steeps, with the sequential extension of C15-C18 leading to the production of the long C56 fatty acid chains, which are precursors of mycolic acids (Marrakchi et al., 2000Marrakchi H, Lanéelle G and Quémard A (2000) InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology (Reading) 146:289-296.; Rawat et al., 2003Ramaswamy S and Musser JM (1998) Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis 79:3-29.; Massengo-Tiassé and Cronan, 2009Massengo-Tiassé RP and Cronan JE (2009) Diversity in enoyl-acyl carrier protein reductases. Cell Mol Life Sci 66:1507-1517.; Zhu et al., 2013Zhu L, Bi H, Ma J, Hu Z, Zhang W, Cronan JE and Wang H (2013) The two functional enoyl-acyl carrier protein reductases of Enterococcus faecalis do not mediate triclosan resistance. mBio 4:e00613-13.; Vögeli et al., 2018Vögeli B, Rosenthal RG, Stoffel GMM, Wagner T, Kiefer P, Cortina NS, Shima S and Erb TJ (2018) InhA, the enoyl-thioester reductase from Mycobacterium tuberculosis forms a covalent adduct during catalysis. J Biol Chem 293:17200-17207.). At the InhA active site, INH-NAD competes with NADH inhibiting its activity and consequently causing the accumulation of saturated C26 fatty acids and stopping the production of mycolic acids, which are key components of the mycobacterial cell wall, contributing to the bacteria lysis (Wilming and Johnsson, 1999Wilming M and Johnsson K (1999) Spontaneous formation of the bioactive form of the tuberculosis drug isoniazid. Angew Chemie Int Ed Engl 38:2588-2590.; Rawat et al., 2003Rawat R, Whitty A and Tonge PJ (2003) The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: Adduct affinity and drug resistance. Proc Natl Acad Sci U S A 100:13881-13886.).
InhA inhibition by INH-NAD adduct. Mechanism of action of InhA and inhibition by INH. KatG activates INH to produce the adduct INH-NAD adduct, which is formed through a reaction with NADH. InhA is inhibited by INH-NAD adduct, which blocks the fatty acid elongation catalyzed by the FAS-II system. Adapted from Vilchèze and Jacobs 2007Vilchèze C and Jacobs WR (2007) The mechanism of isoniazid killing: Clarity through the scope of genetics. Annu Rev Microbiol 61:35-50..
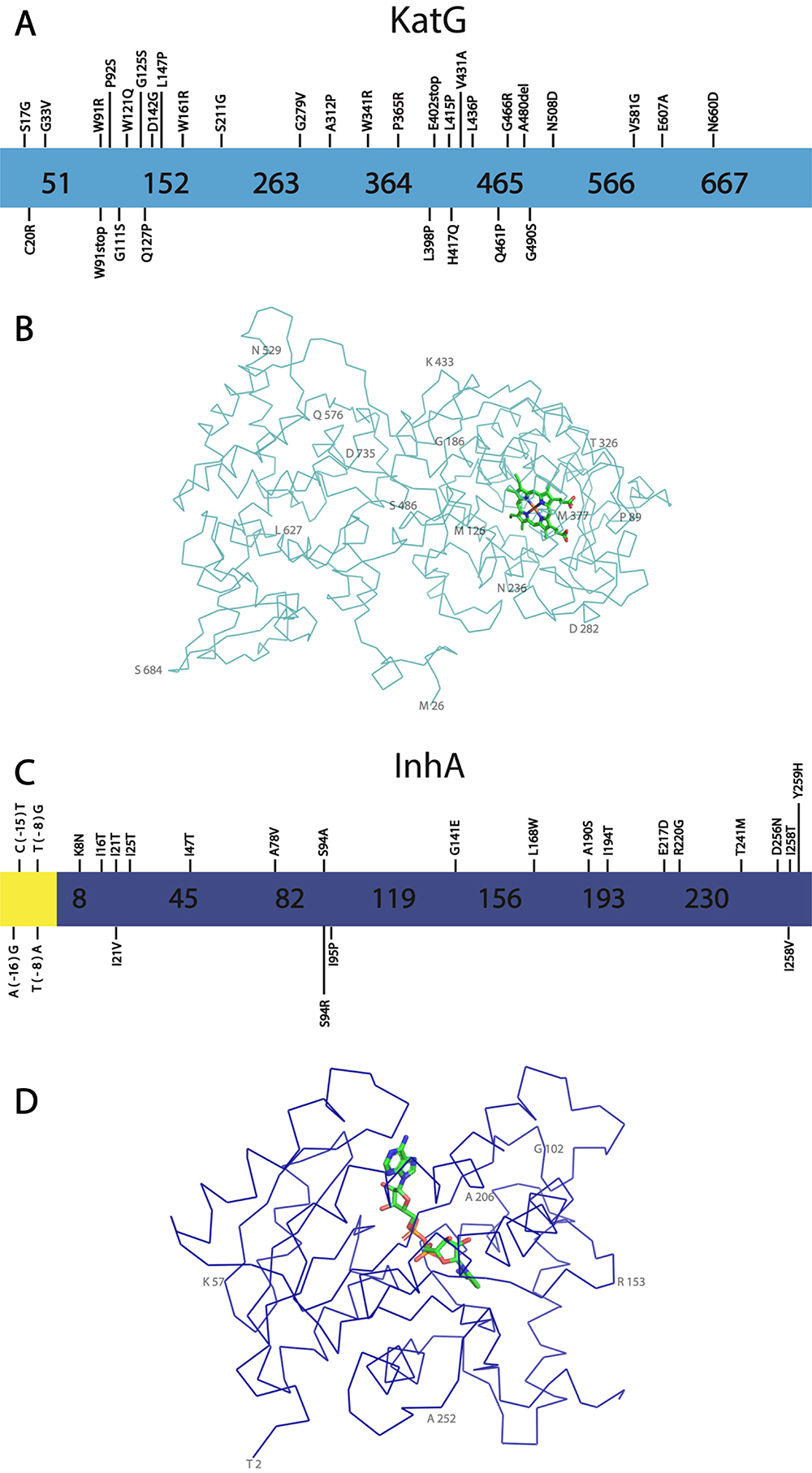
Structure and key mutations for KatG and InhA, proteins involved in INH resistance. A - Representation of KatG primary sequence is shown light blue, showing novel mutations. B - Representation of KatG structure. The contours of Cα are shown in light blue, while the Heme group is shown with carbon atoms in light green. Some residues are numbered in 50% opacity for better visualization. The KatG structure was obtained from PDB entry 2CCA (Zhao et al., 2006Zhao X, Yu H, Yu S, Wang F, Sacchettini JC and Magliozzo RS (2006) Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry 45:4131-4140.) and visualized using the software PyMOL (Schrödinger, 2015Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, Morris SL and Jacobs WR (2002) A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med 8:1171-1174.). C - Representation of InhA sequence in dark blue, showing the most significant mutations. The InhA promoter region is shown in yellow. D - Representation of the structure. The contours of Cα are shown in dark blue while the NADH is shown with carbon atoms in light green. Some residues are numbered in 50% opacity for better visualization. The structure was obtained from PDB entry 4TRN (Chollet et al., 2015Chollet A, Mourey L, Lherbet C, Delbot A, Julien S, Baltas M, Bernadou J, Pratviel G, Maveyraud L and Bernardes-Génisson V (2015) Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach. J Struct Biol 190:328-337.) and the figure was prepared using the software PyMOL (Schrödinger, 2015Schrödinger LLC (2015) The {PyMOL} Molecular Graphics System. Version 1.8. ).
The INH mechanism of resistance is complex and not completely understood and maybe involves many genes, although reported mutations are predominantly identified on katG and inhA genes (Zhang et al., 1992Zhang Y, Heym B, Allen B, Young D and Cole S (1992) The catalase - Peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593.). The mutations observed in inhA and KatG are mostly single missense point substitutions rather than deletions. However, deleterious mutations can also be found on katG (Heym et al., 1999Heym B, Saint-Joanis B and Cole ST (1999) The molecular basis of isoniazid resistance in Mycobacterium tuberculosis. Tuber Lung Dis 79:267-271.) since this gene is a non-essential for Mtb survival. Indeed, the disruption of this gene brings an adaptative advantage under INH treatment and consequently contributes to spreading the INH resistance (Wengenack et al., 2004Wengenack NL, Lane BD, Hill PJ, Uhl JR, Lukat-Rodgers GS, Hall L, Roberts GD, Cockerill FR, Brennan PJ, Rodgers KR et al. (2004) Purification and characterization of Mycobacterium tuberculosis KatG, KatG(S315T), and Mycobacterium bovis KatG(R463L). Protein Expr Purif 36:232-243.).
KatG is a homodimeric enzyme, in which each monomer has 2 domains. These domains have a similar folding to other proteins from the peroxidase family, which are predominantly formed by α-helices (Bertrand et al., 2004Bertrand T, Eady NAJ, Jones JN, Jesmin I, Nagy JM, Jamart-Grégoire B, Raven EL and Brown KA (2004) Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991-38999.). Although, both C-terminal and N-terminal domains are similar, the N-terminal domain binds a heme porphyrin, which is also part of the KatG active site, and the binding of INH closer to the heme-binding site was shown to be a key prerequisite for INH activation (Figure 4B) (Zhao et al., 2006Zhao X, Yu H, Yu S, Wang F, Sacchettini JC and Magliozzo RS (2006) Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry 45:4131-4140.).
In terms of INH resistance, the most common substitutions on KatG occur in the active site, particularly at the INH binding site, which includes the residues R104, H108 and S315. Additionally, several substitutions are also observed in residues involved in the heme binding site (Figure 4B), such as H270 and T275 (Ramaswamy and Musser, 1998Plinke C, Cox HS, Zarkua N, Karimovich HA, Braker K, Diel R, Rüsch-Gerdes S, Feuerriegel S and Niemann S (2010) embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother 65:1359-1367.). Mutations in these residues alter substrate affinity, or change the accessibility to the heme group, leading to lower catalase-peroxidase activity, and causing inefficiency in the INH activation (Carpena et al., 2003Carpena X, Loprasert S, Mongkolsuk S, Switala J, Loewen PC and Fita I (2003) Catalase-peroxidase KatG of Burkholderia pseudomallei at 1.7 Å resolution. J Mol Biol 327:475-489.; Bertrand et al., 2004Bertrand T, Eady NAJ, Jones JN, Jesmin I, Nagy JM, Jamart-Grégoire B, Raven EL and Brown KA (2004) Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991-38999.).
InhA mutations also represent an important factor for INH resistance. One of the most common mechanisms of INH resistance involves the overexpression of inhA gene, caused mainly by mutations in the promoter region (Musser et al., 1996Ohno H, Koga H, Kohno S, Tashiro T and Hara K (1996) Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother 40:1053-1056.; Ramaswamy and Musser, 1998Plinke C, Cox HS, Zarkua N, Karimovich HA, Braker K, Diel R, Rüsch-Gerdes S, Feuerriegel S and Niemann S (2010) embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother 65:1359-1367.; Vilchèze et al., 2006Vilchèze C and Jacobs WRJ (2014) Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: Genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013.). Alternatively, several mutants were also identified to have substitutions on the coding region of inhA leading to missense mutations. Most of the mutant enzymes that were biochemically or biophysically characterized so far indicate a decrease in the NADH binding affinity to InhA and consequently to INH-NAD, increasing the turnover of the coenzyme and adduct in the protein active site, which favors the enzymatic catalysis (Banerjee et al., 1994Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, De Lisle G and Jacobs WR (1994) inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230.; Whitney and Wainberg, 2002Whitney JB and Wainberg MA (2002) Isoniazid, the frontline of resistance in Mycobacterium tuberculosis. McGill J Med 6:114-123.).
A review published by Unissa et al. (2016Unissa AN, Subbian S, Hanna LE and Selvakumar N (2016) Overview on mechanisms of isoniazid action and resistance in Mycobacterium tuberculosis. Infect Genet Evol 45:474-492.) describes in detail several KatG and InhA mutations discovered up to 2016, and from those, the most clinically relevant are briefly reported herein, along with more recently described mutations (Unissa et al., 2016Unissa AN, Subbian S, Hanna LE and Selvakumar N (2016) Overview on mechanisms of isoniazid action and resistance in Mycobacterium tuberculosis. Infect Genet Evol 45:474-492.).
The S315T mutation in KatG is one of the most predominant in INH resistant (INHR) Mtb strains. This substitution leads to the narrowing of the access pathway to the heme group from 6Å to 4.7Å, which decreases the binding affinity for INH and reduces the INH activation and NAD-INH adduct formation. Interestingly, this substitution partially maintains the KatG catalase-peroxidase function (Yu et al., 2003Yu S, Girotto S, Lee C and Magliozzo RS (2003) Reduced affinity for isoniazid in the S315T mutant of Mycobacterium tuberculosis KatG is a key factor in antibiotic resistance. J Biol Chem 278:14769-14775.; Ghiladi et al., 2004Ghiladi RA, Cabelli DE and Ortiz De Montellano PR (2004) Superoxide reactivity of KatG: insights into isoniazid resistance pathways in TB. J Am Chem Soc 126:4772-4773.; Zhao et al., 2006Zhao X, Yu H, Yu S, Wang F, Sacchettini JC and Magliozzo RS (2006) Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry 45:4131-4140.).
On the other hand, His108 is also reported to be a residue involved in INH binding. Two substitutions that have been identified include H108E and H108Q. These two mutations also decrease the affinity of KatG to INH, probably because of weaker interaction and charge repulsion with the substituted residue to the INH hydrazinyl group, which disturbs the INH activation pathway (Musser et al., 1996Ohno H, Koga H, Kohno S, Tashiro T and Hara K (1996) Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother 40:1053-1056.; Bertrand et al., 2004Bertrand T, Eady NAJ, Jones JN, Jesmin I, Nagy JM, Jamart-Grégoire B, Raven EL and Brown KA (2004) Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991-38999.).
A110V mutant has also been reported to cause resistance to INH. The larger side chain of valine alters the H108 conformation, leading to an inefficiency in the binding of INH while also maintaining its enzymatic activity (Wei et al., 2003Wei CJ, Lei B, Musser JM and Tu SC (2003) Isoniazid activation defects in recombinant Mycobacterium tuberculosis catalase-peroxidase (KatG) mutants evident in InhA inhibitor production. Antimicrob Agents Chemother 47:670-675.). Other mutations such as substitution T275P lead to protein instability, which produces an unfolded protein, which is neither capable of activating INH nor catalyzing its reaction (Saint-Joanis et al., 1999Safi H, Sayers B, Hazbón MH and Alland D (2008) Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob Agents Chemother 52:2027-2034.).
Since 2017, several new single nucleotide polymorphisms (SNP) for KatG which are associated with INH resistance have been isolated, and some are summarized herein (Table S1 Table S1 - Novel KatG mutations. ) (Figure 4A). Thwe et al. (2021Thwe EP, Namwat W, Pinlaor P, Rueangsak K and Sangka A (2021) Novel mutations detected from drug resistant Mycobacterium tuberculosis isolated from North East of Thailand. World J Microbiol Biotechnol 37:194.) performed a study using PCR and DNA sequencing in 65 drug-resistant Mtb isolates, revealed a number of new mutations on katG (Thwe et al., 2021). A novel single nucleotide polymorphism (SNP) that causes INH resistance was identified in two of the studied isolates. This mutation is the substitution of proline for arginine at position 365 (P365R) (Thwe et al., 2021Thwe EP, Namwat W, Pinlaor P, Rueangsak K and Sangka A (2021) Novel mutations detected from drug resistant Mycobacterium tuberculosis isolated from North East of Thailand. World J Microbiol Biotechnol 37:194.). This mutation was not characterized, however, although the residue P365 is more than 16Å away from the KatG active site, it was assumed to cause INH resistance. The substitution of proline for arginine probably causes conformational changes that lead to a displacement of key secondary structure elements leading to a lower binding affinity for INH. In this study, only KatG has been evaluated and other mutations in different genes could also be present difficulting the categorization of this mutation as the unique mechanism of resistance to INH.
Kandler et al. (2018Kadura S, King N, Nakhoul M, Zhu H, Theron G, Köser CU and Farhat M (2020) Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 75:2031-2043.), also analyzed 52 INHR Mtb strains using whole-genome sequencing (WGS) and identified 5 novel mutations in KatG, including those that lead to residues substitutions W121Q, W161R, E402stop, A480del, L415P (Kandler et al., 2018Kadura S, King N, Nakhoul M, Zhu H, Theron G, Köser CU and Farhat M (2020) Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 75:2031-2043.). Islam et al. (2019Islam MM, Tan Y, Hameed HMA, Liu Z, Chhotaray C, Liu Y, Lu Z, Cai X, Tang Y, Gao Y et al. (2019) Detection of novel mutations associated with independent resistance and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect 25:1041.e1-1041.e7) performed a large-scale susceptibility test using 10 drugs, including INH, RIF and EMB, against 206 clinical isolates. For KatG, several novel mutations involved in INH resistance were identified, particularly C20R, G33V, W91stop, W91R, P92S, G111S, G125S, Q127P, D142G, L147P, S211G, G279V, A312P, H417Q, V431A, L436P, Q461P, G466R, G490S, V581G, N508D, E607A and N660D, (Islam et al.2019Islam MM, Tan Y, Hameed HMA, Liu Z, Chhotaray C, Liu Y, Lu Z, Cai X, Tang Y, Gao Y et al. (2019) Detection of novel mutations associated with independent resistance and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect 25:1041.e1-1041.e7). Interestingly, the mutations G111S (7.9 Å), Q127P (12.5 Å), D142G (10.7 Å), G279V (10.3 Å) and A312P (8.0 Å) are the closest to the heme group binding cavity (Islam et al., 2019Islam MM, Tan Y, Hameed HMA, Liu Z, Chhotaray C, Liu Y, Lu Z, Cai X, Tang Y, Gao Y et al. (2019) Detection of novel mutations associated with independent resistance and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect 25:1041.e1-1041.e7) and should have an impact in INH activation. These mutations have not been validated through genetic and functional experiments, but may represent great prospects for INH resistance markers.
WGS was also used for the drug resistance prediction in 137 drug-resistant Mtb isolates from Shanghai and 78 from Russia. This study also identified a novel KatG mutation, S17G, which has relevance to INH resistance (Wang et al., 2022Wang L, Yang J, Chen L, Wang W, Yu F and Xiong H (2022) Whole-genome sequencing of Mycobacterium tuberculosis for prediction of drug resistance. Epidemiol Infect 150:e22. ). The N-terminal region of KatG seems to be involved in inter-domain interactions, which is important to dimerization (Bertrand et al., 2004Bertrand T, Eady NAJ, Jones JN, Jesmin I, Nagy JM, Jamart-Grégoire B, Raven EL and Brown KA (2004) Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991-38999.). The mutations S17G, C20R and G33V may cause instability of the protein dimerization. Residues 278 to 312 have been reported to be part of a loop that is possibly involved in the INH binding site (Bertrand et al., 2004Bertrand T, Eady NAJ, Jones JN, Jesmin I, Nagy JM, Jamart-Grégoire B, Raven EL and Brown KA (2004) Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991-38999.) and then, the mutation G279V should also be involved in decreasing the INH binding affinity but not altering the KatG function. More studies are necessary to categorize those novel mutations as resistant conferring and to determine the resistance mechanism, although the WGS strategy has had success in predicting resistant-conferring mutations in Mtb (Kandler et al., 2018Kadura S, King N, Nakhoul M, Zhu H, Theron G, Köser CU and Farhat M (2020) Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 75:2031-2043.).
Alongside katG mutations, other mutations often found and reported to confer INH resistance are those in the inhA promoter region, such as T(− 8)G/A, C(− 15)T and A(− 16)G. These mutations cause overexpression of InhA rather than structural protein modifications (Musser et al., 1996Ohno H, Koga H, Kohno S, Tashiro T and Hara K (1996) Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother 40:1053-1056.; Ramaswamy and Musser, 1998Plinke C, Cox HS, Zarkua N, Karimovich HA, Braker K, Diel R, Rüsch-Gerdes S, Feuerriegel S and Niemann S (2010) embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother 65:1359-1367.; Vilchèze et al., 2006Vilchèze C and Jacobs WRJ (2014) Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: Genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013.) (Figure 4C). According to a data compilation of inhA mutations performed by Seifert et al. (2015Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M and Zhang Y (1997) Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 41:540-543.), in which they analyzed more than 11000 isolates, mutations in inhA promoter region, particularly -15 and -8 were observed in approximately 20,5% of the 6,192 phenotypically resistant isolates, while amino acid substitutions on inhA coding region were found in only approximately 1% of the analyzed strains (Seifert et al., 2015Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M and Zhang Y (1997) Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 41:540-543.). Those mutations involved in the coding region and that are clinically relevant include K8N, I16T, I21T, I25T, I47T, A78V, S94A, S94R, I95P, L168W, A190S, I194T, R202G, E217D, T241M, D256N, I258T, I258V and Y259H (Figure 4C) (Vilchèze et al., 2006Vilchèze C and Jacobs WRJ (2014) Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: Genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013.; Vilchèze and Jacobs, 2014Vilchèze C, Wang F, Arai M, Hazbón MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC and Jacobs WR (2006) Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med 12:1027-1029.; Seifert et al., 2015Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M and Zhang Y (1997) Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 41:540-543.). Missense mutations affecting inhA usually cause an influx of water molecules into the INH-NAD binding site, which decreases NAD-INH adduct and NADH binding affinity. Due to the lower binding affinity, the enzyme turnover rate is altered, changing the bound time of the coenzyme and adduct in the InhA active site. This effect increases the proportion between unbound InhA and bound InhA, and INH-NAD and NADH renovation rate will be higher for the mutants compared to the WT InhA. As consequence, the higher turnover guarantees certain activity of the enzyme even in the presence of INH-NAD (Dessen et al., 1995Dessen A, Quémard A, Blanchard JS, Jacobs WR and Sacchettini JC (1995) Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 267:1638-1641.; Oliveira et al., 2006Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893-900.; Vilchèze et al., 2006Vilchèze C and Jacobs WRJ (2014) Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: Genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013.; Dias et al., 2007Dias MVB, Vasconcelos IB, Prado AMX, Fadel V, Basso LA, de Azevedo WF and Santos DS (2007) Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis. J Struct Biol 159:369-380.). Several mutations, including I21T, I47T, S94A and I95P were biochemically or biophysically characterized. For most of these mutant proteins, it has been observed that the Kd (dissociation constant) for NADH is much higher than for the wild-type enzyme. Interestingly, the mutant I95P did not show any activity in vitro, which indicates that the protein-protein complex formation of the enzymes from the FAS-II system could be important for its residual activity (Basso et al., 1998Basso LA, Zheng R, Musser JM, Jacobs WR and Blanchard JS (1998) Mechanisms of isoniazid resistance in Mycobacterium tuberculosis: Enzymatic characterization of enoyl reductase mutants identified in isoniazid-resistant clinical isolates. J Infect Dis 178:769-775.). The substitution S94A is the most structurally and functionally characterized. Ser94 hydroxyl side chain performs an indirect hydrogen bond with NADH through a water molecule. In the mutated protein, this water molecule is lacking because of the apolar side chain of alanine. As a consequence, there is a direct impact on the affinity of the coenzyme to InhA (Dessen et al., 1995Dessen A, Quémard A, Blanchard JS, Jacobs WR and Sacchettini JC (1995) Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 267:1638-1641.; Oliveira et al., 2006Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893-900.; Vilchèze et al., 2006Vilchèze C and Jacobs WRJ (2014) Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: Genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013.; Dias et al., 2007Dias MVB, Vasconcelos IB, Prado AMX, Fadel V, Basso LA, de Azevedo WF and Santos DS (2007) Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis. J Struct Biol 159:369-380.). On the other hand, Oliveira et al., 2006Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893-900. and Dias et al., 2007Dias MVB, Vasconcelos IB, Prado AMX, Fadel V, Basso LA, de Azevedo WF and Santos DS (2007) Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis. J Struct Biol 159:369-380. further biochemically or structurally investigated the mutations I21V and I47T and observed an altered NADH dissociation constant and a perturbation in water molecules in the active site of the mutant enzymes in comparison to the wild-type enzyme (Oliveira et al., 2006Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893-900.; Dias et al., 2007Dias MVB, Vasconcelos IB, Prado AMX, Fadel V, Basso LA, de Azevedo WF and Santos DS (2007) Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis. J Struct Biol 159:369-380.). Although the performed biophysical studies in a few InhA mutants have shown similar molecular resistance mechanisms, others should exist since there are mutations covering the whole extension of the InhA primary structure (Figure 4C) (Vilchèze et al., 2006Vilchèze C and Jacobs WRJ (2014) Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: Genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013.; Vilchèze and Jacobs, 2014Vilchèze C, Wang F, Arai M, Hazbón MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC and Jacobs WR (2006) Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med 12:1027-1029.; Seifert et al., 2015Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M and Zhang Y (1997) Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 41:540-543.). In addition, novel mutations in the inhA gene have continually appeared, such as the recent identification of substitution G141E, which has been identified in a resistant Mtb isolate harboring KatG mutation D142G and EthA S266R, but has not been validated yet as resistant-conferring through genetic and functional assays. (Islam et al., 2019Islam MM, Tan Y, Hameed HMA, Liu Z, Chhotaray C, Liu Y, Lu Z, Cai X, Tang Y, Gao Y et al. (2019) Detection of novel mutations associated with independent resistance and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect 25:1041.e1-1041.e7).
Resistance to RIF
Rifampicin (RIF), also known as Rifampin, is an antimicrobial chemically derived from the rifamycins, which are members of the ansamycins antibiotic family. Rifamycin compounds consist of 7 different molecules, in which RIF is a derivative of Rifamycin SV. Rifamycin SV chemical structure was altered by the synthetical addition of a 3-(4-methyl-1-piperazinyl)-iminomethyl group, improving chemical stability and oral administration while maintaining high antibacterial activity (Murray et al., 2015Musser JM, Kapur V, Williams DL, Kreiswirth BN, Van Soolingen D and Van Embden JDA (1996) Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: Restricted array of mutations associated with drug resistance. J Infect Dis 173:196-202.; Brucoli and McHugh, 2021Brucoli F and McHugh TD (2021) Rifamycins: Do not throw the baby out with the bathwater. Is rifampicin still an effective anti-tuberculosis drug? Future Med Chem 13:2129-2131.; Zloh et al., 2021Zloh M, Gupta M, Parish T and Brucoli F (2021) Novel C-3-(N-alkyl-aryl)-aminomethyl rifamycin SV derivatives exhibit activity against rifampicin-resistant Mycobacterium tuberculosis RpoBS522L strain and display a different binding mode at the RNAP β-subunit site compared to rifampicin. Eur J Med Chem 225:113734.).
RIF was first synthesized in 1965 and since 1970 has been used for TB treatment, particularly in combined therapy with INH (Grobbelaar et al., 2019Grobbelaar M, Louw GE, Sampson SL, van Helden PD, Donald PR and Warren RM (2019) Evolution of rifampicin treatment for tuberculosis. Infect Genet Evol 74:103937.). The addition of RIF to the treatment regimen together with INH and EMB also enabled the therapy to be shortened from 12 to 9 months because of its higher efficacy in sterilization. Thus, currently, RIF is considered a first-line drug against TB, and part of the standard drug combination used for TB treatment (Murray et al., 2015Musser JM, Kapur V, Williams DL, Kreiswirth BN, Van Soolingen D and Van Embden JDA (1996) Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: Restricted array of mutations associated with drug resistance. J Infect Dis 173:196-202.; Grobbelaar et al., 2019Grobbelaar M, Louw GE, Sampson SL, van Helden PD, Donald PR and Warren RM (2019) Evolution of rifampicin treatment for tuberculosis. Infect Genet Evol 74:103937.; World Health Organization, 2021World Health Organization (2021) Global Tuberculosis Report 2021. ).
RIF inhibits RNA synthesis by binding to the DNA-dependent -RNA polymerase (RNAP) (McClure and Cech, 1978McClure WR and Cech CL (1978) On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem 253:8949-8956.). RIF binds to the β-subunit of RNAP, encoded by the rpoB gene, and causes a steric clash between the 5’ phosphates elongating RNA. Thus, RIF inhibits the RNA elongation path for RNA transcripts of 2 or 3 nucleotides in length during the translocation movement of RNAP. This blockage severely disturbs the bacterial transcription mechanism and consequently leads to cell death (McClure and Cech, 1978McCune RM and Tompsett R (1956) Fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. I. The persistence of drug-susceptible tubercle bacilli in the tissues despite prolonged antimicrobial therapy. J Exp Med 104:737-762.; Campbell et al., 2001Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A and Darst SA (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912.; Zloh et al., 2021Zloh M, Gupta M, Parish T and Brucoli F (2021) Novel C-3-(N-alkyl-aryl)-aminomethyl rifamycin SV derivatives exhibit activity against rifampicin-resistant Mycobacterium tuberculosis RpoBS522L strain and display a different binding mode at the RNAP β-subunit site compared to rifampicin. Eur J Med Chem 225:113734.).
RIF resistance is mainly associated with mutations in the rpoB gene, which encodes the subunit β of RNAP (Figure 5). RNAP is an essential protein, and its sequence is extremely conserved in all bacteria (Campbell et al., 2001Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A and Darst SA (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912.; Betts et al., 2002Betts JC, Lukey PT, Robb LC, McAdam RA and Duncan K (2002) Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717-731.; Goldstein, 2014Goldstein BP (2014) Resistance to rifampicin: A review. J Antibiot (Tokyo) 67:625-630.). It has been extensively reported that the region between codons 426 to 452 in Mtb and 507 to 533 in E.coli of the rpoB gene is more commonly affected by mutations and these are often involved in RIF resistance. This region contains 81 bp and is usually referred to as R-resistance determining region (RRDR). The amino acid chain encoded by RRDR was confirmed to be important to RIF binding and consequently, mutations in this region affect the affinity of RIF to RNAP (Campbell et al., 2001Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A and Darst SA (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912.; Pang et al., 2013Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893-900.; Goldstein, 2014Goldstein BP (2014) Resistance to rifampicin: A review. J Antibiot (Tokyo) 67:625-630.). Thus, about 95% of all RIF-resistant (RIFR) strains have mutations in this region, particularly at codons 516, 526, and 531 (435, 445, and 450 in Mtb), which are responsible for nearly 90% of all known RIFR strains (Figure 5) (Ohno et al., 1996Oliveira JS, Pereira JH, Canduri F, Rodrigues NC, de Souza ON, de Azevedo WF, Basso LA and Santos DS (2006) Crystallographic and pre-steady-state kinetics studies on binding of NADH to wild-type and isoniazid-resistant enoyl-ACP(CoA) reductase enzymes from Mycobacterium tuberculosis. J Mol Biol 359:646-666.; Somoskovi et al., 2001Smith T, Wolff KA and Nguyen L (2012) Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr Top Microbiol Immunol 374:53-80; Campbell et al., 2001Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A and Darst SA (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912.; Goldstein, 2014Goldstein BP (2014) Resistance to rifampicin: A review. J Antibiot (Tokyo) 67:625-630.).
Structure and key mutations for β-subunit of RNAP encoded by rpoB gene, the main target of RIF and the most reported protein in RIF resistance. A - Representation of β-subunit of RNAP primary sequence in orange, showing the most relevant mutations. Substitutions are shown in black, while insertions and deletions are shown in red. B - The β-subunit of RNAP structure. The representation in orange shows the Cα contours while RIF is shown with carbon atoms in light green. The β-subunit of RNAPR encoded by rpoB was obtained from PDB entry 5UHD (Lin et al., 2017Lin W, Mandal S, Degen D, Liu Y, Ebright YW, Li S, Feng Y, Zhang Y, Mandal S, Jiang Y et al. (2017) Structural basis of Mycobacterium tuberculosis transcription and transcription inhibition. Mol Cell 66:169-179.e8.) and the figure was prepared using the software PyMOL (Schrödinger, 2015Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, Morris SL and Jacobs WR (2002) A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med 8:1171-1174.). Some residues are numbered in 50% opacity for better visualization.
This mechanism of resistance can be exemplified by the substitutions S531L, which is the most common substitution or by H526D (Figure 5). Both of these mutations interfere with intramolecular hydrogen bonds. The side chain of the mutant L531 disrupts the RIF binding because of a structural clash with RIF binding position, according to Pang et al., 2013Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893-900.. On the other hand, D526 causes a charge repulsion decreasing the RIF binding affinity (Mcnerney et al., 2000Mikhailovich V, Lapa S, Gryadunov D, Sobolev A, Strizhkov B, Chernyh N, Skotnikova O, Irtuganova O, Moroz A, Litvinov V et al. (2001) Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips. J Clin Microbiol 39:2531-2540.; Mikhailovich et al., 2001Miotto P, Cabibbe AM, Borroni E, Degano M and Cirilloa DM (2018) Role of disputed mutations in the rpoB gene in interpretation of automated liquid MGIT culture results for rifampin susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol 56:e01599-17.; Pang et al., 2013Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893-900.). Other substitutions that are also involved in RIF resistance, but usually have a higher phenotypic variation, including those showing a lower level of resistance, are H526L, H526G, H526R and L533P (Mcnerney et al., 2000Mikhailovich V, Lapa S, Gryadunov D, Sobolev A, Strizhkov B, Chernyh N, Skotnikova O, Irtuganova O, Moroz A, Litvinov V et al. (2001) Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips. J Clin Microbiol 39:2531-2540.; Mikhailovich et al., 2001Miotto P, Cabibbe AM, Borroni E, Degano M and Cirilloa DM (2018) Role of disputed mutations in the rpoB gene in interpretation of automated liquid MGIT culture results for rifampin susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol 56:e01599-17.; Pang et al., 2013Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893-900.). In addition, Hirani et al. 2020Hirani N, Joshi A, Anand S, Chowdhary A, Ganesan K, Agarwal M and Phadke N (2020) Detection of a novel mutation in the rpoB gene in a multidrug resistant Mycobacterium tuberculosis isolate using whole genome next generation sequencing. J Glob Antimicrob Resist 22:270-274. also identified from an MDR-TB isolate from India, a rpoB mutation that encodes an insertion of an arginine between the residues 512 and 513 (512-Arg-514) (Hirani et al., 2020Hirani N, Joshi A, Anand S, Chowdhary A, Ganesan K, Agarwal M and Phadke N (2020) Detection of a novel mutation in the rpoB gene in a multidrug resistant Mycobacterium tuberculosis isolate using whole genome next generation sequencing. J Glob Antimicrob Resist 22:270-274.). Interestingly, other insertions in the RRDR region are also commonly reported and associated with RIF resistance, particularly 514 L 516, 511 P 513, 511 E 513 (Sinha et al., 2020Shoeb HA, Bowman BU, Ottolenghi AC and Merola AJ (1985) Evidence for the generation of active oxygen by isoniazid treatment of extracts of Mycobacterium tuberculosis H37Ra. Antimicrob Agents Chemother 27:404-407.) (Figure 5A).
A number of substitutions have also been reported to change other residues outside the RRDR. In the last 5 years, RIFR strains have been continually analyzed, and many new rpoB mutations were identified (Table S2 Table S2 - Novel RpoB mutations. ). Takawira et al. (2017Sun Q, Li X, Perez LM, Shi W, Zhang Y and Sacchettini JC (2020) The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD. Nat Commun 11:339.) collected 100 isolates from Zimbabwe, and detected 13 novel mutations, varying from substitution mutations inside RRDR, such as R529Q, and also outside of RRDR, such as I572P that have not yet been validated as resistant-conferring mutations but have been identified in resistant strains. (Takawira et al., 2017Takawira FT, Mandishora RSD, Dhlamini Z, Munemo E and Stray-Pedersen B (2017) Mutations in rpoB and katG genes of multidrug resistant Mycobacterium tuberculosis undetectable using genotyping diagnostic methods. Pan Afr Med J 27:145.) (Figure 5). Maningi et al. (2018Maningi NE, Daum LT, Rodriguez JD, Said HM, Peters RPH, Sekyere JO, Fischer GW, Chambers JP and Fouriea PB (2018) Multi- and extensively drug resistant Mycobacterium tuberculosis in South Africa: A molecular analysis of historical isolates. J Clin Microbiol 56:e01214-17.) also conducted a study using 240 isolates from South Africa and identified 5 mutations outside of the RRDR, of which 2 of them are associated with RIF resistance, T480A, and Q253R (Maningi et al., 2018Maningi NE, Daum LT, Rodriguez JD, Said HM, Peters RPH, Sekyere JO, Fischer GW, Chambers JP and Fouriea PB (2018) Multi- and extensively drug resistant Mycobacterium tuberculosis in South Africa: A molecular analysis of historical isolates. J Clin Microbiol 56:e01214-17.). Finally, more recently, a WGS was used for the prediction of drug resistance in 137 drug-resistant Mtb isolates from Shanghai and 78 from Russia. This study has identified a novel rpoB mutation that encodes rpoB with a substitution Q172R (Wang et al., 2022Wang L, Yang J, Chen L, Wang W, Yu F and Xiong H (2022) Whole-genome sequencing of Mycobacterium tuberculosis for prediction of drug resistance. Epidemiol Infect 150:e22. ). Although more studies are necessary to categorize this novel mutation as resistant conferring and to determine the resistance mechanism, the WGS strategy has had success in predicting resistant-conferring mutations in Mtb (Kandler et al., 2018Kadura S, King N, Nakhoul M, Zhu H, Theron G, Köser CU and Farhat M (2020) Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 75:2031-2043.).
Additionally, approximately 5% of all (RIFR) strains do not have mutations in rpoB gene at all. Several studies propose that the mechanism of RIF could be the overexpression of genes involved in bacterial efflux mechanisms, such as mmr, mmpL7, Rv1258c, p55 and efpA. This hypothesis indicates that resistance in Mtb can emerge from a combination of specific mutations and bacterial metabolic adaptations (Machado et al., 2017Machado D, Coelho TS, Perdigão J, Pereira C, Couto I, Portugal I, Maschmann RDA, Ramos DF, von Groll A, Rossetti MLR et al. (2017) Interplay between mutations and efflux in drug resistant clinical isolates of Mycobacterium tuberculosis. Front Microbiol 8:711.).
In the context of drug susceptibility tests (DST) for Mtb, it is important to highlight that although phenotypic DST methods are the “gold standard” for the detection of Rifampicin resistance-conferring mutations, there are some mutations that are consistently missed by those methods, which is the case for mutations conferring low-level RIF resistance, commonly referred as disputed mutations. These mutations, which include L511P and D516Y, are associated with poor clinical outcomes but evade the detection of phenotypic DST screenings. In such circumstances, genotypic DST, such as WGS, is promising for the detection of disputed mutations. Genotypic DST can also provide important information regarding compensatory mutations that restore bacterial fitness due to a decrease in this characteristic caused by mutations in the RRDR of rpoB. These compensatory mutations were reported outside of RRDR in rpoB, or even in other genes, such as rpoA/C, and can directly impact the clinical outcome for such strains (Ahmad and Mokaddas, 2014Ahmad S and Mokaddas E (2014) Current status and future trends in the diagnosis and treatment of drug-susceptible and multidrug-resistant tuberculosis. J Infect Public Health 7:75-91.; Miotto et al., 2018Mirnejad R, Asadi A, Khoshnood S, Mirzaei H, Heidary M, Fattorini L, Ghodousi A and Darban-Sarokhalil D (2018) Clofazimine: A useful antibiotic for drug-resistant tuberculosis. Biomed Pharmacother 105:1353-1359.; Al-Mutairi et al., 2019aAl-Mutairi NM, Ahmad S, Mokaddas E, Eldeen HS and Joseph S (2019a) Occurrence of disputed rpoB mutations among Mycobacterium tuberculosis isolates phenotypically susceptible to rifampicin in a country with a low incidence of multidrug-resistant tuberculosis. BMC Infect Dis 19:3.,bAl-Mutairi NM, Ahmad S and Mokaddas EM (2019b) Molecular characterization of multidrug-resistant Mycobacterium tuberculosis (MDR-TB) isolates identifies local transmission of infection in Kuwait, a country with a low incidence of TB and MDR-TB. Eur J Med Res 24:38.; Ma et al., 2021Ma P, Luo T, Ge L, Chen Z, Wang X, Zhao R, Liao W and Bao L (2021) Compensatory effects of M. tuberculosis rpoB mutations outside the rifampicin resistance-determining region. Emerg Microbes Infect 10:743-752.; Shea et al., 2021Selikoff IJ, Robitzek EH and Ornstein GG (1952) Treatment of pulmonary tuberculosis with hydrazide derivatives of isonicotinic acid. J Am Med Assoc 150:973-980.).
Resistance to PZA
Pyrazinamide (PZA) is an analog of nicotinamide, with the substitution of the pyridine group for pyrazine. This drug was first synthesized in 1936, but it was only acknowledged as a potential drug for the treatment of TB in 1952. Since 1970, PZA has been used as a first-line drug for TB treatment. PZA is the unique anti-TB drug that is selective against latent TB and the addition of this drug to the previous TB treatment regimen, composed of INH, RIF and EMB, was essential for the TB treatment regimen reduction from 9 to 6 months (Fox et al., 1999Fox W, Ellard GA and Mitchison DA (1999) Studies on the treatment of tuberculosis undertaken by the British Medical Research Council Tuberculosis Units, 1946-1986, with relevant subsequent publications. Int J Tuberc Lung Dis 3:S231-279.; Murray et al., 2015Musser JM, Kapur V, Williams DL, Kreiswirth BN, Van Soolingen D and Van Embden JDA (1996) Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: Restricted array of mutations associated with drug resistance. J Infect Dis 173:196-202.).
Pyrazinamide (PZA) is an analog of nicotinamide, with the substitution of the pyridine group for pyrazine. This drug was first synthesized in 1936, but it was only acknowledged as a potential drug for the treatment of TB in 1952. Since 1970, PZA has been used as a first-line drug for TB treatment. PZA is the unique anti-TB drug that is selective against latent TB and the addition of this drug to the previous TB treatment regimen, composed of INH, RIF and EMB, was essential for the TB treatment regimen reduction from 9 to 6 months (Fox et al., 1999Fox W, Ellard GA and Mitchison DA (1999) Studies on the treatment of tuberculosis undertaken by the British Medical Research Council Tuberculosis Units, 1946-1986, with relevant subsequent publications. Int J Tuberc Lung Dis 3:S231-279.; Murray et al., 2015Musser JM, Kapur V, Williams DL, Kreiswirth BN, Van Soolingen D and Van Embden JDA (1996) Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: Restricted array of mutations associated with drug resistance. J Infect Dis 173:196-202.).
PZA, similarly to INH, is also a prodrug and needs to be converted to pyrazinoic acid (POA) by the nicotinamidase or pyrazinamidase (PZAse) (Figure 6), encoded by the pncA gene, to exhibit its antitubercular activity (Scorpio and Zhang, 1996Schrödinger LLC (2015) The {PyMOL} Molecular Graphics System. Version 1.8. ). PZA/POA shows lesser activity in growing bacteria, and greater, in the persistent or latent stage, having an important role as a sterilizing drug (Mccune and Tompsett, 1956McNerney R, Kiepiela P, Bishop KS, Nye PM and Stoker NG (2000) Rapid screening of Mycobacterium tuberculosis for susceptibility to rifampicin and streptomycin. Int J Tuberc Lung Dis 4:69-75.; Mitchison, 1985Murray JF, Schraufnagel DE and Hopewell PC (2015) Treatment of tuberculosis: A historical perspective. Ann Am Thorac Soc 12:1749-1759.; Hu et al., 2006Hu Y, Coates AR and Mitchison DA (2006) Sterilising action of pyrazinamide in models of dormant and rifampicin-tolerant Mycobacterium tuberculosis. Int J Tuberc Lung Dis 10:317-322.).
Structure and relevant mutations for PncA and PanD, enzymes involved in PZA resistance. A - Representation of the PncA primary sequence in light green, showing key mutations. Substitutions are shown in black, while insertions and deletions are sown in red. The pncA promoter region is shown in yellow. B - Structure of PncA. The contours of Cα are shown in light green. PncA structure was obtained from PDB entry 3PL1 (Petrella et al., 2011Petrella S, Gelus-Ziental N, Maudry A, Laurans C, Boudjelloul R and Sougakoff W (2011) Crystal structure of the pyrazinamidase of Mycobacterium tuberculosis: Insights into natural and acquired resistance to pyrazinamide. PLoS One 6:e15785.) and the figure was prepared using the software PyMOL (Schrödinger, 2015Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, Morris SL and Jacobs WR (2002) A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med 8:1171-1174.). Some residues are numbered in 50% opacity for better visualization. C - Representation of the PanD primary sequence is shown in dark green, highlighting the most significant mutations. D - PanD Structure. The contours of Cα are shown in dark green, while PZA is shown with carbon atoms in light green. The PanD Structure was obtained from PDB entry 6OZ8 (Sun et al., 2020Sun Q, Xiao TY, Liu HC, Zhao XQ, Liu ZG, Li YN, Zeng H, Zhao LL and Wan KL (2018) Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17.) and the figure was prepared using the software PyMOL (Schrödinger, 2015). A number of residues are numbered in 50% opacity for better visualization.
The most accepted mechanism of action for this drug is that in acidic environments, such as inflamed tissues as a consequence of Mtb activity, the POA influx into the bacterial cell is facilitated, leading to POA accumulation inside the bacillus and the eventual cytoplasm acidification (Zhang et al., 2014Zhang Y, Shi W, Zhang W and Mitchison D (2014) Mechanisms of Pyrazinamide Action and Resistance. Microbiol Spectr. 2:MGM2-0023-2013.). POA also seems capable of disrupting membrane potential and of de-energizing the membrane (Zhang et al., 1999Zhang Y, Scorpio A, Nikaido H and Sun Z (1999) Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Bacteriol 181:2044-2049., 2003Zhang Y, Shi W, Zhang W and Mitchison D (2014) Mechanisms of Pyrazinamide Action and Resistance. Microbiol Spectr. 2:MGM2-0023-2013.). POA may also be involved in the inhibition of the trans-translation process through the binding to protein RpsA (Shi et al., 2011Shea J, Halse TA, Kohlerschmidt D, Lapierre P, Modestil AHA, Kearns CH, Dworkin FF, Rakeman JL, Escuyer V and Musser KA (2021) Low-level rifampin resistance and rpoB mutations in Mycobacterium tuberculosis: An analysis of whole-genome sequencing and drug susceptibility test data in new york. J Clin Microbiol 59:e01885-20.).
In recent years, strong evidence indicated that the enzyme PanD is one of the major targets for PZA/POA (Sun et al., 2020Sun Q, Xiao TY, Liu HC, Zhao XQ, Liu ZG, Li YN, Zeng H, Zhao LL and Wan KL (2018) Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17.). PanD (Figure 6) is an enzyme involved in the CoA and pantothenate biosynthesis, being directly involved in β-alanine production (Zhang et al., 2003Zhang Y, Wade MM, Scorpio A, Zhang H and Sun Z (2003) Mode of action of pyrazinamide: Disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother 52:790-795., 2014Zhang Y, Shi W, Zhang W and Mitchison D (2014) Mechanisms of Pyrazinamide Action and Resistance. Microbiol Spectr. 2:MGM2-0023-2013.). CoA and pantothenate are key molecules to Mtb persistence, which may explain why PZA/POA is more active for bacteria in these conditions (Zhang et al., 2002Zhang Y, Permar S and Sun Z (2002) Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol 51:42-49.; Sambandamurthy et al., 2002Saint-Joanis B, Souchon H, Wilming M, Johnsson K, Alzari PM and Cole ST (1999) Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem J 338:753-760.). POA binds to the PanD active site and competes with its substrate, D-aspartate. The Mtb PanD crystal structure was solved in complex with POA, which revealed the interactions of this ligand and the protein. Particularly, it was revealed that POA performs key hydrogen interactions with A74, A75, R54 from the PanD active site. (Sun et al., 2020Sun Q, Xiao TY, Liu HC, Zhao XQ, Liu ZG, Li YN, Zeng H, Zhao LL and Wan KL (2018) Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17.).
Mutations that cause resistance to PZA in Mtb have been observed predominantly in pncA gene impacting the PZase activity, leading to a decrease in the efficiency of PZA activation (Konno et al., 1967Kidwai S, Park CY, Mawatwal S, Tiwari P, Jung MG, Gosain TP, Kumar P, Alland D, Kumar S, Bajaj A et al. (2017) Dual mechanism of action of 5-Nitro-1,10-phenanthroline against Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e00969-17.; Petrella et al., 2011Petrella S, Gelus-Ziental N, Maudry A, Laurans C, Boudjelloul R and Sougakoff W (2011) Crystal structure of the pyrazinamidase of Mycobacterium tuberculosis: Insights into natural and acquired resistance to pyrazinamide. PLoS One 6:e15785.) (Figure 6B); however, a number of mutations have also been observed in panD and rpsA genes (Shi et al., 2011Shea J, Halse TA, Kohlerschmidt D, Lapierre P, Modestil AHA, Kearns CH, Dworkin FF, Rakeman JL, Escuyer V and Musser KA (2021) Low-level rifampin resistance and rpoB mutations in Mycobacterium tuberculosis: An analysis of whole-genome sequencing and drug susceptibility test data in new york. J Clin Microbiol 59:e01885-20.; Feuerriegel et al., 2013Feuerriegel S, Köser CU, Richter E and Niemann S (2013) Mycobacterium canettii is intrinsically resistant to both pyrazinamide and pyrazinoic acid. J Antimicrob Chemother 68:1439-1440.; Sun et al., 2020Sun Q, Xiao TY, Liu HC, Zhao XQ, Liu ZG, Li YN, Zeng H, Zhao LL and Wan KL (2018) Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17.). In the case of mutations on PanD, PZA resistance should be caused by a lower POA affinity and decreasing the residence time on PanD active site. Particularly, mutations on PanD residues of two loops that cover the active site have been reported. These PanD loops are formed by residues 20-24 and 119-126, which are involved in the formation of a barrier over the protein active site to maintain the substrate isolated from the solvent (Figure 6D) (Sun et al., 2020Sun Q, Xiao TY, Liu HC, Zhao XQ, Liu ZG, Li YN, Zeng H, Zhao LL and Wan KL (2018) Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17.).
Sun et al. (2020Sun Q, Xiao TY, Liu HC, Zhao XQ, Liu ZG, Li YN, Zeng H, Zhao LL and Wan KL (2018) Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17.) confirmed that the PanD mutations H21R and M117I are associated with PZA resistance. These researchers investigated using isothermal titration calorimetry (ITC) and enzymatic assays the affinity and the activity of these two mutants and they observed higher affinity of POA and stronger inhibition for the wild-type enzyme in contrast to the mutants (Sun et al., 2020Sun Q, Xiao TY, Liu HC, Zhao XQ, Liu ZG, Li YN, Zeng H, Zhao LL and Wan KL (2018) Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17.). Using crystallography, it was also confirmed that the mutations H21R and M117I affect regions that are near the C-terminal loops of the α and β-chains. Still, Hameed et al. (2020Hameed HMA, Tan Y, Islam MM, Lu Z, Chhotaray C, Wang S, Liu Z, Fang C, Tan S, Yew WW et al. (2020) Detection of novel gene mutations associated with pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates in Southern China. Infect Drug Resist 13:217-227.) reported the identification of a novel PanD mutation, L132P, isolated in clinical isolates from Southern China. The mutation L132P is interesting due to its proximity to the PanD C-terminal loop (Figure 6D). However, currently, there are no biochemical or biophysical characterization studies for this mutation (Hameed et al., 2020Hameed HMA, Tan Y, Islam MM, Lu Z, Chhotaray C, Wang S, Liu Z, Fang C, Tan S, Yew WW et al. (2020) Detection of novel gene mutations associated with pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates in Southern China. Infect Drug Resist 13:217-227.). Other clinically important mutations for PanD are H21R, I49V, E130G, P134S, and V138A H21R, I49V, E130G, P134S, and V138A (Zhang et al., 2013Zhang S, Chen J, Shi W, Liu W, Zhang W and Zhang Y (2013) Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect 2:e34.), but there are also no functional studies about the effect of these substitutions on the enzyme and ligand affinities (Figure 6D).
Although mutations can also occur through whole pncA extension, this gene has three major regions that are mostly affected by mutations and polymorphisms which are: Nucleotides 3-17, 61-85 and 132-142 (Scorpio et al., 1997Scorpio A and Zhang Y (1996) Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med 2:662-667.; Zhang et al., 2014Zhang Y, Shi W, Zhang W and Mitchison D (2014) Mechanisms of Pyrazinamide Action and Resistance. Microbiol Spectr. 2:MGM2-0023-2013.). These regions seem to be part of three different loops that are key regions of the active site architecture. Daum et al. (2019Daum LT, Konstantynovska OS, Solodiankin OS, Poteiko PI, Bolotin VI, Rodriguez JD, Gerilovych AP, Chambers JP and Fischer GW (2019) Characterization of novel Mycobacterium tuberculosis pncA gene mutations in clinical isolates from the Ukraine. Diagn Microbiol Infect Dis 93:334-338.) performed a study using 91 Mtb clinical isolates from Ukraine, and identified a number of mutations in pncA gene. 11 novel mutations were identified and those can be divided by substitution mutations: Q10H, V93M, G132R, A146P, T177P; deletion mutations: Promoter Δ(-5), ΔIV(6,7)(deletion of isoleucine and valine at positions six and seven, respectively); frameshift insertions: 4 frameshift (cgTTG), 16 frameshift (GGgT), 132 frameshift (cGGT); and insertions: 122 frameshift (cggCAA). Of these mutations, ΔIV(6,7), Q10H, and V93M were confirmed to cause resistance to PZA (Figure 6B) (Daum et al., 2019Daum LT, Konstantynovska OS, Solodiankin OS, Poteiko PI, Bolotin VI, Rodriguez JD, Gerilovych AP, Chambers JP and Fischer GW (2019) Characterization of novel Mycobacterium tuberculosis pncA gene mutations in clinical isolates from the Ukraine. Diagn Microbiol Infect Dis 93:334-338.).
Since 2017, several new mutations for PncA that cause PZA resistance have been isolated, and some are summarized herein (Table S3 Table S3 - Novel PncA mutations. ). Khan et al. (2018Keam SJ (2019) Pretomanid: First approval. Drugs 79:1797-1803.) biophysically characterized three PncA mutations, including L19K, R140H and E144K. Using molecular dynamics simulations, these authors observed that the RMSD, obtained through superposition, for the mutant protein models seems to be higher than 2Å when compared to the wild-type (WT) position. This suggests that these mutations disturb the protein stability and reduce its activity against PZA strongly contributing to resistance (Khan et al., 2018Keam SJ (2019) Pretomanid: First approval. Drugs 79:1797-1803.). For these mutants, the hydrogen bonding of the PZAse with POA was disturbed in comparison to the wild-type protein, indicating changes in residues that are part of the ligand-binding site (Khan et al., 2018Keam SJ (2019) Pretomanid: First approval. Drugs 79:1797-1803.).
Li et al. (2021Li D, Song Y, Zhang CL, Li X, Xia X and Zhang AM (2017) Screening mutations in drug-resistant Mycobacterium tuberculosis strains in Yunnan, China. J Infect Public Health 10:630-636.) performed a study using 465 clinical isolates, 424 of those being drug-resistant, identified 30 novel mutations involved with PZA resistance (Table S3 Table S3 - Novel PncA mutations. ). Among them, 24 were confirmed to not have Pzase activity, while 6 maintained it. Those first 24 mutations occur in pncA regions involved in PZase activity, while the other 6 mutants have amino acid substitution in other regions. 15 negative Pzase active mutations were tested for PZA susceptibility, and all were classified as resistant, while the two Pzase positive active mutations were susceptible to PZA (Li et al., 2021Li D, Song Y, Zhang CL, Li X, Xia X and Zhang AM (2017) Screening mutations in drug-resistant Mycobacterium tuberculosis strains in Yunnan, China. J Infect Public Health 10:630-636.).
Resistance to EMB
Ethambutol (EMB) was identified as a drug with potential anti-TB activity in 1960, and it has synergy with INH and it is part of the drug combination used in TB standard treatment, which also includes PZA and RIF. EMB has a relatively simple chemical structure, consisting of an ethylenediamine molecule at the center, with the addition of butanol moieties at the end of each side of the carbon chain (Thomas et al., 1961Thomas JP, Baughn CO, Wilkinson RG and She I (1961) A new synthetic compound with antituberculous activity in mice: Ethambutol (dextro-2,2’-(ethylenediimino)-di-l-butanol). Am Rev Respir Dis 83:891-893.; Murray et al., 2015Musser JM, Kapur V, Williams DL, Kreiswirth BN, Van Soolingen D and Van Embden JDA (1996) Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: Restricted array of mutations associated with drug resistance. J Infect Dis 173:196-202.).
The EMB mechanism of action involves the inhibition of arabinose incorporation at the arabinogalactan from the mycobacterial cell wall, leading to the cell accumulation of decaprenyl-phosphate-arabinose that eventually culminates in bacterium cell death (Takayama and Kilburn, 1989Takayama K and Kilburn JO (1989) Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob Agents Chemother 33:1493-1499.; Sreevatsan et al., 1997Sotgiu G, Centis R, D’Ambrosio L and Battista Migliori G (2015) Tuberculosis treatment and drug regimens. Cold Spring Harb Perspect Med 5:a017822.; Telenti et al., 1997Telenti A, Philipp WJ, Sreevatsan S, Bernasconi C, Stockbauer KE, Wieles B, Musser JM and Jacobs WR (1997) The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med 3:567-570.). The operon embCAB encodes the proteins EmbA, EmbB and EmbC, which are arabinosyltransferases involved in the cell wall arabinan biosynthesis (Belanger et al., 1996Belanger AE, Besra GS, Ford ME, Mikušová K, Belisle JT, Brennan PJ and Inamine JM (1996) The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci U S A 93:11919-11924.; Telenti et al., 1997Telenti A, Philipp WJ, Sreevatsan S, Bernasconi C, Stockbauer KE, Wieles B, Musser JM and Jacobs WR (1997) The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med 3:567-570.) (Figure 7). EmbA and EmbB are associated to produce a heterodimeric complex, while EmbC is a homodimeric enzyme (Tan et al., 2020Tan YZ, Rodrigues J, Keener JE, Zheng RB, Brunton R, Kloss B, Giacometti SI, Rosário AL, Zhang L, Niederweis M et al. (2020) Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis. Nat Commun 11:3396.; Zhang et al., 2020Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS et al. (2020) Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368:1211-1219.; Bendre et al., 2021Bendre AD, Peters PJ and Kumar J (2021) Recent insights into the structure and function of mycobacterial membrane proteins facilitated by Cryo-EM. J Membr Biol 254:321-341.). Figure 7B shows the structure of EmbB, which has the most key mutations involved in EMB resistance. The cryo-EM tridimensional structures of these three proteins in complex with their glycosyl donor substrate, decaprenyl-phosphate-arabinose; the acceptor substrate, diarabinose; and the known inhibitor, EMB, have been determined, and it was confirmed that EMB binds competitively to the active sites of EmbB and EmbC (Goude et al., 2009Goude R, Amin AG, Chatterjee D and Parish T (2009) The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis. Antimicrob Agents Chemother 53:4138-4146.; Tan et al., 2020Tan YZ, Rodrigues J, Keener JE, Zheng RB, Brunton R, Kloss B, Giacometti SI, Rosário AL, Zhang L, Niederweis M et al. (2020) Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis. Nat Commun 11:3396.). An acyl carrier protein, AcpM, was also attached to the Emb proteins. The complex EmbA-EmbB-AcpM catalyzes the transfer of an arabinose molecule from the acceptor to the donor producing a 1-3 linkage between the arabinose residue and the arabinan receptor, while the complex EmbC-Acp)M is involved in the extension of the arabinan chain, following a 1-5 linkage (Zhang et al., 2020Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS et al. (2020) Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368:1211-1219.; Bendre et al., 2021Bendre AD, Peters PJ and Kumar J (2021) Recent insights into the structure and function of mycobacterial membrane proteins facilitated by Cryo-EM. J Membr Biol 254:321-341.). EMB was shown to inhibit these reactions occupying the disaccharide product binding site (Wolucka et al., 1994Wolucka BA, McNeil MR, De Hoffmann E, Chojnacki T and Brennan PJ (1994) Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J Biol Chem 269:23328-23335.).
Structure and key mutations involved in the embCAB operon and their proteins, which are involved with EMB resistance. A - Representation of embCAB operon and the position of mutations involved in the resistance to EMB. The EmbC, EmbA and EmbB sequences are represented in purple, red and pink, respectively, showing the mutations described herein. The intergenic spaces between the gene encoding regions are shown in yellow. B - EmbB Structure. The Cα contours are shown in pink. EmbB structure was obtained from PDB entry 7BVF (Zhang et al., 2020Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS et al. (2020) Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368:1211-1219.) and the figure was prepared using the software PyMOL (Schrödinger, 2015Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, Morris SL and Jacobs WR (2002) A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med 8:1171-1174.). A number of residues are numbered in 50% opacity for better visualization.
Additionally, EMB has synergistic activity with INH. It is proposed that EtbR, a transcription factor encoded by the Rv0273c gene, is responsible for negatively regulating InhA expression, acting as a repressor for InhA expression. It was proposed that EMB is capable of interacting with this repressor, EtbR, amplifying its effect. This ultimately would lead to even less InhA expression, and consequently, higher INH susceptibility, in the co-administration with EMB (Zhu et al., 2018Zhu C, Liu Y, Hu L, Yang M and He ZG (2018) Molecular mechanism of the synergistic activity of ethambutol and isoniazid against Mycobacterium tuberculosis. J Biol Chem 293:16741-16750.).
The EMB resistance mostly involves mutations in the embCAB operon, which encodes target proteins of EMB. Even so, in 30% of EMB-resistant (EMBR) strains, mutations in embB gene are not reported (Zhang and Yew, 2015Zhang Y and Yew WW (2015) Mechanisms of drug resistance in Mycobacterium tuberculosis: Update 2015. Int J Tuberc Lung Dis 19:1276-1289. ). Mutations in embC and the embC-embA intergenic space were also reported to EMB resistance (Cui et al., 2014Cui Z, Li Y, Cheng S, Yang H, Lu J, Hu Z and Ge B (2014) Mutations in the embC-embA intergenic region contribute to Mycobacterium tuberculosis resistance to ethambutol. Antimicrob Agents Chemother 58:6837-6843.).
Mutations on the embC gene may lead to the production of mutant proteins with a lower EMB binding affinity due to substitutions near the protein binding site to EMB, while mutations in embC-embA intergenic space may have a strong impact on the mRNA expression of embA and embB genes (Cui et al., 2014Cui Z, Li Y, Cheng S, Yang H, Lu J, Hu Z and Ge B (2014) Mutations in the embC-embA intergenic region contribute to Mycobacterium tuberculosis resistance to ethambutol. Antimicrob Agents Chemother 58:6837-6843.; Zhang et al., 2020Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS et al. (2020) Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368:1211-1219.).
Mutations in the residue M306 from EmbB have been identified in more than 68% of EMBR isolates (Safi et al., 2008Rifat D, Li SY, Ioerger T, Shah K, Lanoix JP, Lee J, Bashiri G, Sacchettini J and Nuermberger E (2021) Mutations in fbiD (Rv2983) as a novel determinant of resistance to pretomanid and delamanid in Mycobacterium tuberculosis. Antimicrob Agents Chemother 65:e01948-20.; Zhang and Yew, 2015Zhang Y and Yew WW (2015) Mechanisms of drug resistance in Mycobacterium tuberculosis: Update 2015. Int J Tuberc Lung Dis 19:1276-1289. ). Furthermore, G406 and Q497 have also been reported as resistance hotspots and mutations in these sites are frequently isolated in EMBR strains (Plinke et al., 2010Plinke C, Cox HS, Zarkua N, Karimovich HA, Braker K, Diel R, Rüsch-Gerdes S, Feuerriegel S and Niemann S (2010) embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother 65:1359-1367.; Dai et al., 2019Dai E, Zhang H, Zhou X, Song Q, Li D, Luo L, Xu X, Jiang W and Ling H (2019) MycoResistance: A curated resource of drug resistance molecules in Mycobacteria. Database (Oxford) 2019:baz074.). Based on the structure of EmbB, M306 is directly involved in EMB binding, and mutations affecting this residue, or those residues that interact with it, including Y302 and E327, disturb the EMB binding affinity because of differences in the interaction between the protein active site and EMB (Plinke et al., 2009Plinke C, Cox HS, Kalon S, Doshetov D, Rüsch-Gerdes S and Niemann S (2009) Tuberculosis ethambutol resistance: Concordance between phenotypic and genotypic test results. Tuberculosis (Edinb) 89:448-452., 2010Plinke C, Cox HS, Zarkua N, Karimovich HA, Braker K, Diel R, Rüsch-Gerdes S, Feuerriegel S and Niemann S (2010) embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother 65:1359-1367.; Zhang et al., 2020Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS et al. (2020) Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368:1211-1219.) (Figure 7). On the other hand, mutations affecting Q497 may interfere with the interaction of E327 and EMB, which decreases the binding affinity of this drug. Finally, the resistance mechanisms involved in mutations affecting G406 are proposed to lead to a steric hindrance and consequently cause conformational changes in EMB binding site, decreasing the ligand affinity (Plinke et al., 2009Plinke C, Cox HS, Kalon S, Doshetov D, Rüsch-Gerdes S and Niemann S (2009) Tuberculosis ethambutol resistance: Concordance between phenotypic and genotypic test results. Tuberculosis (Edinb) 89:448-452., 2010Plinke C, Cox HS, Kalon S, Doshetov D, Rüsch-Gerdes S and Niemann S (2009) Tuberculosis ethambutol resistance: Concordance between phenotypic and genotypic test results. Tuberculosis (Edinb) 89:448-452.; Zhang et al., 2020Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS et al. (2020) Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368:1211-1219.).
Since 2017, a number of mutations in proteins encoded by the embCAB operon have been reported (Table S4 Table S4 - Novel EmbC, EmbA, EmbB mutations. ). Li et al. (2017Lange C, Chesov D and Heyckendorf J (2019) Clofazimine for the treatment of multidrug-resistant tuberculosis. Clin Microbiol Infect 25:128-130.) identified 54 resistant Mtb isolates after performing susceptibility tests from Yunnan, China. These authors identified a novel mutation for embB, which encodes the substitution D78G (Li et al., 2017) (Figure 7A). However, a closer analysis of the position of this residue indicates that it is not involved in the protein active site and it has not been validated through gene replacement phenotypic assays as a resistance-conferring mutation.
Park et al. (2018aPark J, Jang W, Kim M, Kim Y, Shin SY, Park K, Kim MS and Shin S (2018a) Molecular drug resistance profiles of Mycobacterium tuberculosis from sputum specimens using ion semiconductor sequencing. J Microbiol Methods 145:1-6.) analyzed 34 resistant isolates from Daejeon City in South Korea and reported two novel EmbB mutations, S317P and Q445R (Park et al., 2018aPark J, Jang W, Kim M, Kim Y, Shin SY, Park K, Kim MS and Shin S (2018a) Molecular drug resistance profiles of Mycobacterium tuberculosis from sputum specimens using ion semiconductor sequencing. J Microbiol Methods 145:1-6.). Still, Park et al. (2018b) also identified two novel embB mutations from 30 MDR-TB isolates from Cheongju, Korea, that encodes the substitutions of Y319D and H1002R (Park et al., 2018bPark J, Shin SY, Kim K, Park K, Shin S and Ihm C (2018b) Determining genotypic drug resistance by ion semiconductor sequencing with the ion ampliSeqTM TB panel in multidrug-resistant Mycobacterium tuberculosis isolates. Ann Lab Med 38:316-323.). Those mutations have been correlated with EMB resistance since phenotypical DST of the isolates showed resistance, but were not yet validated through gene replacement (Figure 7A).
Sun et al. (2018Stancil SL, Mirzayev F and Abdel-Rahman SM (2021) Profiling pretomanid as a therapeutic option for TB infection: Evidence to date. Drug Des Devel Ther 15:2815-2830.) tested 125 isolates from China and measured the MIC of these isolates against EMB. The authors identified 2 novel mutations for gene embA: one which is located in the intergenic region, embAG(−5)A, and the other one that encodes the substitution V18F (Figure 7A). These authors also identified a novel mutation in embC that encodes the substitution D329E (Sun et al., 2018Stancil SL, Mirzayev F and Abdel-Rahman SM (2021) Profiling pretomanid as a therapeutic option for TB infection: Evidence to date. Drug Des Devel Ther 15:2815-2830.). However, the mutations embAG(−5)A and V18F were identified in isolates that also have other embB mutations, which are well known to be involved in EMB resistance (Figure 7A). Although mutations in the intergenic region of embA-embC are known to upregulate the expression of proteins encoded by embCAB operon, there is no validation whether these related mutations are solely involved in EMB resistance (Cui et al., 2014Cui Z, Li Y, Cheng S, Yang H, Lu J, Hu Z and Ge B (2014) Mutations in the embC-embA intergenic region contribute to Mycobacterium tuberculosis resistance to ethambutol. Antimicrob Agents Chemother 58:6837-6843.; Sun et al., 2018Stancil SL, Mirzayev F and Abdel-Rahman SM (2021) Profiling pretomanid as a therapeutic option for TB infection: Evidence to date. Drug Des Devel Ther 15:2815-2830.).
In the context of DST for EMB, it has been reported that phenotypic DST methods have a higher percentage of false susceptibility results for EMBr strains than genotypical and molecular DST methods. The lower sensitivity of the phenotypical DSTs is mainly explained due to the slower activity of EMB for Mtb, and the even lower activity of the drug in the culture media used, which explains EMBr strains harboring mutations in codons 306 or 406 categorized as susceptible, while it is highly unlikely that those mutations are not associated with EMB resistance (Ahmad et al., 2007Ahmad S, Jaber AA and Mokaddas E (2007) Frequency of embB codon 306 mutations in ethambutol-susceptible and -resistant clinical Mycobacterium tuberculosis isolates in Kuwait. Tuberculosis (Edinb) 87:123-129.; Ahmad et al., 2016Ahmad S, Mokaddas E, Al-Mutairi N, Eldeen HS and Mohammadi S (2016) Discordance across phenotypic and molecular methods for drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates in a low TB incidence country. PLoS One 11:e0153563.; Al-Mutairi et al., 2019bAl-Mutairi NM, Ahmad S and Mokaddas EM (2019b) Molecular characterization of multidrug-resistant Mycobacterium tuberculosis (MDR-TB) isolates identifies local transmission of infection in Kuwait, a country with a low incidence of TB and MDR-TB. Eur J Med Res 24:38.).
Resistance to other drugs
Bedaquiline (BDQ) was one of the last approved drugs by the FDA to combat TB after a void of about 30 years. This drug has antimycobacterial activity based on the inhibition of mycobacteria ATP synthase without affecting the human enzyme. BDQ is a diarylquinolone, structurally organized with amine and alcohol side chains (Patel et al., 2019Patel H, Pawara R, Pawara K, Ahmed F, Shirkhedkar A and Surana S (2019) A structural insight of bedaquiline for the cardiotoxicity and hepatotoxicity. Tuberculosis (Edinb) 117:79-84.; Khoshnood et al., 2021Khan MT, Junaid M, Mao X, Wang Y, Hussain A, Malik SI and Wei DQ (2018) Pyrazinamide resistance and mutations L19R, R140H, and E144K in Pyrazinamidase of Mycobacterium tuberculosis. J Cell Biochem 120:7154-7166.). Canonically, ATP synthase is formed by a membrane domain, F0, with the subunits a, b2 and c10, and by a cytoplasmatic domain, F1, which is constituted by the subunits a3, b3, g, d and ε (Khoshnood et al., 2021Khan MT, Junaid M, Mao X, Wang Y, Hussain A, Malik SI and Wei DQ (2018) Pyrazinamide resistance and mutations L19R, R140H, and E144K in Pyrazinamidase of Mycobacterium tuberculosis. J Cell Biochem 120:7154-7166.). BDQ inhibits ATP synthase by the binding in the F0 domain of the Mtb ATP synthase, particularly in the subunit c and subunits a-c interface, specifically interacting with the residues L63, E65, A66, A67 Y68 and I70 from c subunit and L70, P172 and I173 from the a subunit (Guo et al., 2021Guo H, Courbon GM, Bueler SA, Mai J, Liu J and Rubinstein JL (2021) Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. Nature 589:143-147.).
BDQ resistance has already been observed and it is commonly associated with mutations in specific genes, including atpE, which encodes the subunit c of ATP synthase and the transcription regulation factor gene mmpR (Li et al., 2019Li Y, Sun F and Zhang W (2019) Bedaquiline and delamanid in the treatment of multidrug-resistant tuberculosis: Promising but challenging. Drug Dev Res 80:98-105.; Kadura et al., 2020Ismail N, Rivière E, Limberis J, Huo S, Metcalfe JZ, Warren RM and Van Rie A (2021) Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2:e604-e616.; Battaglia et al., 2020Battaglia S, Spitaleri A, Cabibbe AM, Meehan CJ, Utpatel C, Ismail N, Tahseen S, Skrahina A, Alikhanova N, Mostofa Kamal SM et al. (2020) Characterization of genomic variants associated with resistance to bedaquiline and delamanid in naive Mycobacterium tuberculosis clinical strains. J Clin Microbiol 58:1304-1324.; Goossens et al., 2021Goossens SN, Sampson SL and Van Rie A (2021) Mechanisms of drug-induced tolerance in Mycobacterium tuberculosis. Clin Microbiol Rev 34:e00141-20.). These mutations include D28V, E61D, A63P and I66M in ATP synthase subunit c, and mutations W42R, and S53L for MmpR. It has also been observed frameshifts in the region between the codons 38-144 for the mmpR gene (Kadura et al., 2020Ismail N, Rivière E, Limberis J, Huo S, Metcalfe JZ, Warren RM and Van Rie A (2021) Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2:e604-e616.). Nevertheless, Ismail et al. (2021Ismail N, Rivière E, Limberis J, Huo S, Metcalfe JZ, Warren RM and Van Rie A (2021) Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2:e604-e616.) summarized 14 atpE and 237 mmpR polymorphisms reported in the literature between 2006 to 2020. From these, many mutations were associated with BDQ resistance, including atpE mutations that were reported in clinical BDQ-resistant strains. However, further confirmations of such polymorphisms are those solely responsible for the BDQ resistance still need to be confirmed. (Ismail et al., 2021Ismail N, Rivière E, Limberis J, Huo S, Metcalfe JZ, Warren RM and Van Rie A (2021) Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2:e604-e616.).
Pretomanid (Pa) and Delamanid (DLM) are nitroimidazole antimycobacterial agents, with a nitroimidazooxazine core. Pa is currently used in association with BDQ and Linezolid (LZD) against MDR-TB. Pa and DLM mechanism of action involves the inhibition of mycobacterial mycolic acids and cell wall biosynthesis. Nitroimidazoles are usually activated by the enzyme Ddn, which binds the unusual co-factor F420. Pa and DLM showed antimycobacterial activity against replicating and non-replicating bacteria (Li et al., 2019Li K, Yang Z, Gu J, Luo M, Deng J and Chen Y (2021) Characterization of pncA Mutations and Prediction of PZA Resistance in Mycobacterium tuberculosis Clinical Isolates From Chongqing, China. Front Microbiol 11:594171.). One of the proposed mechanisms of action for Pa and DLM in replicating bacteria is the oxidation of precursors necessary for lipid biosynthesis, inhibiting this process. For non-replicating bacteria, followed by the Ddn activation of Pa/DLM, these drugs might generate reactive nitrogen species, such as nitric oxide that can interact with cellular components and lead to bacterial death (Keam, 2019Kandler JL, Mercante AD, Dalton TL, Ezewudo MN, Cowan LS, Burns SP, Metchock B, Cegielski P, Posey JE and Global PETTS Investigators (2018) Validation of novel Mycobacterium tuberculosis isoniazid resistance mutations not detectable by common molecular tests. Antimicrob Agents Chemother 62:e00974-18.; Stancil et al., 2021Sreevatsan S, Stockbauer KE, Pan X, Kreiswirth BN, Moghazeh SL, Jacobs WR, Telenti A and Musser JM (1997) Ethambutol resistance in Mycobacterium tuberculosis: Critical role of embB mutations. Antimicrob Agents Chemother 41:1677-1681.; Hori et al., 2022Hori T, Owusu YB and Sun D (2022) US FDA-Approved Antibiotics During the 21st Century. In: Rezaei N (ed) Encyclopedia of Infection and Immunity, Elsevier, pp 556-585.).
In mutant Mtb strains, Pa and DLM capacity for generating reactive substances is inefficient (Guglielmetti et al., 2020Guglielmetti L, Chiesi S, Eimer J, Dominguez J, Masini T, Varaine F, Veziris N, Ader F and Robert J (2020) Bedaquiline and delamanid for drug-resistant tuberculosis: A clinician’s perspective. Future Microbiol 15:779-799.; Stancil et al., 2021Sreevatsan S, Stockbauer KE, Pan X, Kreiswirth BN, Moghazeh SL, Jacobs WR, Telenti A and Musser JM (1997) Ethambutol resistance in Mycobacterium tuberculosis: Critical role of embB mutations. Antimicrob Agents Chemother 41:1677-1681.). Therefore, the more commonly reported resistance mechanism associated with Pa and DLM involves mutations to key enzymes responsible for nitroimidazole activation, including Ddn and the further enzymes Fgd1, FbiA, FbiB, FbiC and FbiD (Parveen et al., 2021Parveen U, Sultana S, Heba SF, Rafi R, Begum A and Fatima N (2021) Pretomanid: A novel therapeutic paradigm for treatment of drug resistant tuberculosis. Indian J Tuberc 68:106-113.). Several groups have already described a number of mutations in the genes that encode these proteins, including Ddn S11stop and Y133D; fbiC V720I and P372S and fbiD A198P for Pa and Ddn D241A, G242A, W88stop and L107P for DLM (Pang et al., 2017Pang Y, Zong Z, Huo F, Jing W, Ma Y, Dong L, Li Y, Zhao L, Fu Y and Huang H (2017) In vitro drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing, China. Antimicrob Agents Chemother 61:e00900-17.; Kidwai et al., 2017Khosravi AD, Tabandeh MR, Shahi F and Salmanzadeh S (2021) Linezolid resistance among multidrug-resistant Mycobacterium tuberculosis clinical isolates in Iran. Acta Microbiol Immunol Hung 68:203-207.; Yang et al., 2018Yang JS, Kim KJ, Choi H and Lee SH (2018) Delamanid, bedaquiline, and linezolid minimum inhibitory concentration distributions and resistance-related gene mutations in multidrug-resistant and extensively drug-resistant tuberculosis in Korea. Ann Lab Med 38:563-568.; Kadura et al., 2020Ismail N, Rivière E, Limberis J, Huo S, Metcalfe JZ, Warren RM and Van Rie A (2021) Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2:e604-e616.; Rifat et al., 2021Rawat R, Whitty A and Tonge PJ (2003) The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: Adduct affinity and drug resistance. Proc Natl Acad Sci U S A 100:13881-13886.).
Linezolid (LZD) is an oxazolidinone drug with antimicrobial activity by the inhibition of protein synthesis, particularly binding to rRNA 23S from ribosomal subunit 50S. LZD inhibits the formation of the ribosomal complex 70S, leading to a non-functional initiation complex, and, consequently, decreasing the efficiency of the translation process (Hashemian et al., 2018Hashemian SMR, Farhadi T and Ganjparvar M (2018) Linezolid: A review of its properties, function, and use in critical care. Drug Des Devel Ther 12:1759-1767.; Fermeli et al., 2020Fermeli DD, Marantos TD, Liarakos ALD, Panayiotakopoulos GD, Dedes VK and Panoutsopoulos GI (2020) Linezolid: A promising agent for the treatment of multiple and extensively drug-resistant tuberculosis. Folia Med (Plovdiv) 62:444-452.). Structurally, LZD is formed by an N-aryl ring core, which is important to its function as an antimicrobial. LZD was clinically introduced in 1996, and in the last 10 years, its activity against Mtb has been evaluated. Usually, this drug has been repurposed for anti- TB treatment, particularly in cases of MDR-TB and XDR-TB. LZD also seems to possess anti- TB activity against persistent bacteria (Drusano et al., 2018Drusano GL, Myrick J, Maynard M, Nole J, Duncanson B, Brown D, Schmidt S, Neely M, Scanga CA, Peloquin C et al. (2018) Linezolid kills acid-phase and nonreplicative-persister-phase Mycobacterium tuberculosis in a hollow-fiber infection model. Antimicrob Agents Chemother 62:e00221-18.).
The resistance mechanism for LNZ involves, primarily, mutations to ribosomal protein L3 and subunit 23S, encoded by genes rplC and rrl, respectively. The mutation C154R in RplC is one of the most prevalent ones (Ushtanit et al., 2021Ushtanit A, Mikhailova Y, Lyubimova A, Makarova M, Safonova S, Filippov A, Borisov S and Zimenkov D (2021) Genetic profile of linezolid-resistant M. Tuberculosis clinical strains from moscow. Antibiotics (Basel) 10:1243.). Nevertheless, other mutations reported to LZD were summarized by Kadura et al. (2020Ismail N, Rivière E, Limberis J, Huo S, Metcalfe JZ, Warren RM and Van Rie A (2021) Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2:e604-e616.) and Khosravi et al. (2021Khoshnood S, Goudarzi M, Taki E, Darbandi A, Kouhsari E, Heidary M, Motahar M, Moradi M and Bazyar H (2021) Bedaquiline: Current status and future perspectives. J Glob Antimicrob Resist 25:48-59.). The other prevalent mutations include G2270T, G2299, G2746A, G2814T and C2848A for Rrl. These authors also identified 2 polymorphisms in LZD resistance strains for rplC, V141I and I150N. However, they have not been completely validated to be the unique reason for LZD resistance (Kadura et al., 2020; Khosravi et al., 2021).
Clofazimine (CFZ) is a drug repurposed for resistant TB treatment. Structurally, this molecule is a riminophenazine, constituted of a phenazine core with an R-imino group. The mechanism of action of this molecule is not entirely understood, as the envelope, the respiratory chain, or even the DNA were reported as possible targets (Mirnejad et al., 2018Mitchison DA (1985) The action of antituberculosis drugs in short-course chemotherapy. Tubercle 66:219-225.; Lange et al., 2019Konno K, Feldmann FM and McDermott W (1967) Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis 95:461-469.).
The most important mechanism of resistance associated with CFZ includes mutations in the gene mmpR which encodes a transcript regulator of efflux pumps. The most prevalent substitutions include R156*, G193del, and G193Ins (Kadura et al., 2020Ismail N, Rivière E, Limberis J, Huo S, Metcalfe JZ, Warren RM and Van Rie A (2021) Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2:e604-e616.).
Conclusions
In summary, this review compiles the molecular mechanisms of resistance and also describes a number of novel mutations identified in the last five years for the first-line drugs (INH, PZA, RIF and EMB) that are currently used for TB treatment regimens. Identifying, reporting, and understanding the resistance of Mtb to these antimicrobials has pivotal importance because of the difficulty in the management and treatment of TB, particularly those caused by MDR and XDR strains. With the rise of novel methods of genome sequencing and the development of structural biology, particularly cryo-EM, the identification and understanding of the mechanism of resistance has considerably evolved in the last five years allowing the anticipation of the mutational effect. Overall, all advances made in this field might considerably contribute to more personalized use of anti-TB drugs, avoid the dissemination of resistant strains, better management of the antimicrobials used in TB treatment, and the discovery of new medicines.
Acknowledgements
This work was supported by FAPESP, grant numbers 2020/03850-9 and 2022/12234-5. NOR was funded by the FAPESP fellowship (19/17037-0) and MVBD also received a CNPq research productivity fellowship (208998/2020-0).
References
- Ahmad S, Jaber AA and Mokaddas E (2007) Frequency of embB codon 306 mutations in ethambutol-susceptible and -resistant clinical Mycobacterium tuberculosis isolates in Kuwait. Tuberculosis (Edinb) 87:123-129.
- Ahmad S and Mokaddas E (2014) Current status and future trends in the diagnosis and treatment of drug-susceptible and multidrug-resistant tuberculosis. J Infect Public Health 7:75-91.
- Ahmad S, Mokaddas E, Al-Mutairi N, Eldeen HS and Mohammadi S (2016) Discordance across phenotypic and molecular methods for drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates in a low TB incidence country. PLoS One 11:e0153563.
- Al-Mutairi NM, Ahmad S, Mokaddas E, Eldeen HS and Joseph S (2019a) Occurrence of disputed rpoB mutations among Mycobacterium tuberculosis isolates phenotypically susceptible to rifampicin in a country with a low incidence of multidrug-resistant tuberculosis. BMC Infect Dis 19:3.
- Al-Mutairi NM, Ahmad S and Mokaddas EM (2019b) Molecular characterization of multidrug-resistant Mycobacterium tuberculosis (MDR-TB) isolates identifies local transmission of infection in Kuwait, a country with a low incidence of TB and MDR-TB. Eur J Med Res 24:38.
- Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, De Lisle G and Jacobs WR (1994) inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis Science 263:227-230.
- Basso LA, Zheng R, Musser JM, Jacobs WR and Blanchard JS (1998) Mechanisms of isoniazid resistance in Mycobacterium tuberculosis: Enzymatic characterization of enoyl reductase mutants identified in isoniazid-resistant clinical isolates. J Infect Dis 178:769-775.
- Battaglia S, Spitaleri A, Cabibbe AM, Meehan CJ, Utpatel C, Ismail N, Tahseen S, Skrahina A, Alikhanova N, Mostofa Kamal SM et al (2020) Characterization of genomic variants associated with resistance to bedaquiline and delamanid in naive Mycobacterium tuberculosis clinical strains. J Clin Microbiol 58:1304-1324.
- Belanger AE, Besra GS, Ford ME, Mikušová K, Belisle JT, Brennan PJ and Inamine JM (1996) The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci U S A 93:11919-11924.
- Bendre AD, Peters PJ and Kumar J (2021) Recent insights into the structure and function of mycobacterial membrane proteins facilitated by Cryo-EM. J Membr Biol 254:321-341.
- Bertrand T, Eady NAJ, Jones JN, Jesmin I, Nagy JM, Jamart-Grégoire B, Raven EL and Brown KA (2004) Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J Biol Chem 279:38991-38999.
- Betts JC, Lukey PT, Robb LC, McAdam RA and Duncan K (2002) Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717-731.
- Bodiguel J, Nagy JM, Brown KA and Jamart-Grégoire B (2001) Oxidation of isoniazid by manganese and Mycobacterium tuberculosis catalase-peroxidase yields a new mechanism of activation. J Am Chem Soc 123:3832-3833.
- Brucoli F and McHugh TD (2021) Rifamycins: Do not throw the baby out with the bathwater. Is rifampicin still an effective anti-tuberculosis drug? Future Med Chem 13:2129-2131.
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A and Darst SA (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912.
- Carpena X, Loprasert S, Mongkolsuk S, Switala J, Loewen PC and Fita I (2003) Catalase-peroxidase KatG of Burkholderia pseudomallei at 1.7 Å resolution. J Mol Biol 327:475-489.
- Cherinka B, Andrews BH, Sánchez-Gallego J, Brownstein J, Argudo-Fernández M, Blanton M, Bundy K, Jones A, Masters K, Law DR et al (2019) Marvin: A tool kit for streamlined access and visualization of the SDSS-IV MaNGA data set. Astron J 158:74.
- Chollet A, Mourey L, Lherbet C, Delbot A, Julien S, Baltas M, Bernadou J, Pratviel G, Maveyraud L and Bernardes-Génisson V (2015) Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach. J Struct Biol 190:328-337.
- Cui Z, Li Y, Cheng S, Yang H, Lu J, Hu Z and Ge B (2014) Mutations in the embC-embA intergenic region contribute to Mycobacterium tuberculosis resistance to ethambutol. Antimicrob Agents Chemother 58:6837-6843.
- Dai E, Zhang H, Zhou X, Song Q, Li D, Luo L, Xu X, Jiang W and Ling H (2019) MycoResistance: A curated resource of drug resistance molecules in Mycobacteria. Database (Oxford) 2019:baz074.
- Daum LT, Konstantynovska OS, Solodiankin OS, Poteiko PI, Bolotin VI, Rodriguez JD, Gerilovych AP, Chambers JP and Fischer GW (2019) Characterization of novel Mycobacterium tuberculosis pncA gene mutations in clinical isolates from the Ukraine. Diagn Microbiol Infect Dis 93:334-338.
- Dessen A, Quémard A, Blanchard JS, Jacobs WR and Sacchettini JC (1995) Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis Science 267:1638-1641.
- Dias MVB, Vasconcelos IB, Prado AMX, Fadel V, Basso LA, de Azevedo WF and Santos DS (2007) Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis J Struct Biol 159:369-380.
- Drusano GL, Myrick J, Maynard M, Nole J, Duncanson B, Brown D, Schmidt S, Neely M, Scanga CA, Peloquin C et al (2018) Linezolid kills acid-phase and nonreplicative-persister-phase Mycobacterium tuberculosis in a hollow-fiber infection model. Antimicrob Agents Chemother 62:e00221-18.
- Fermeli DD, Marantos TD, Liarakos ALD, Panayiotakopoulos GD, Dedes VK and Panoutsopoulos GI (2020) Linezolid: A promising agent for the treatment of multiple and extensively drug-resistant tuberculosis. Folia Med (Plovdiv) 62:444-452.
- Fernandes GF dos S, Salgado HRN and Santos JL dos (2017) Isoniazid: A review of characteristics, properties and analytical methods. Crit Rev Anal Chem 47:298-308.
- Feuerriegel S, Köser CU, Richter E and Niemann S (2013) Mycobacterium canettii is intrinsically resistant to both pyrazinamide and pyrazinoic acid. J Antimicrob Chemother 68:1439-1440.
- Fox W, Ellard GA and Mitchison DA (1999) Studies on the treatment of tuberculosis undertaken by the British Medical Research Council Tuberculosis Units, 1946-1986, with relevant subsequent publications. Int J Tuberc Lung Dis 3:S231-279.
- Ghiladi RA, Cabelli DE and Ortiz De Montellano PR (2004) Superoxide reactivity of KatG: insights into isoniazid resistance pathways in TB. J Am Chem Soc 126:4772-4773.
- Goldstein BP (2014) Resistance to rifampicin: A review. J Antibiot (Tokyo) 67:625-630.
- Goossens SN, Sampson SL and Van Rie A (2021) Mechanisms of drug-induced tolerance in Mycobacterium tuberculosis Clin Microbiol Rev 34:e00141-20.
- Gopal P and Dick T (2014) Reactive dirty fragments: Implications for tuberculosis drug discovery. Curr Opin Microbiol 21:7-12.
- Goude R, Amin AG, Chatterjee D and Parish T (2009) The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis Antimicrob Agents Chemother 53:4138-4146.
- Grobbelaar M, Louw GE, Sampson SL, van Helden PD, Donald PR and Warren RM (2019) Evolution of rifampicin treatment for tuberculosis. Infect Genet Evol 74:103937.
- Guglielmetti L, Chiesi S, Eimer J, Dominguez J, Masini T, Varaine F, Veziris N, Ader F and Robert J (2020) Bedaquiline and delamanid for drug-resistant tuberculosis: A clinician’s perspective. Future Microbiol 15:779-799.
- Guo H, Courbon GM, Bueler SA, Mai J, Liu J and Rubinstein JL (2021) Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. Nature 589:143-147.
- Hameed HMA, Tan Y, Islam MM, Lu Z, Chhotaray C, Wang S, Liu Z, Fang C, Tan S, Yew WW et al (2020) Detection of novel gene mutations associated with pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates in Southern China. Infect Drug Resist 13:217-227.
- Hashemian SMR, Farhadi T and Ganjparvar M (2018) Linezolid: A review of its properties, function, and use in critical care. Drug Des Devel Ther 12:1759-1767.
- Heym B, Saint-Joanis B and Cole ST (1999) The molecular basis of isoniazid resistance in Mycobacterium tuberculosis Tuber Lung Dis 79:267-271.
- Hirani N, Joshi A, Anand S, Chowdhary A, Ganesan K, Agarwal M and Phadke N (2020) Detection of a novel mutation in the rpoB gene in a multidrug resistant Mycobacterium tuberculosis isolate using whole genome next generation sequencing. J Glob Antimicrob Resist 22:270-274.
- Hori T, Owusu YB and Sun D (2022) US FDA-Approved Antibiotics During the 21st Century. In: Rezaei N (ed) Encyclopedia of Infection and Immunity, Elsevier, pp 556-585.
- Hu Y, Coates AR and Mitchison DA (2006) Sterilising action of pyrazinamide in models of dormant and rifampicin-tolerant Mycobacterium tuberculosis Int J Tuberc Lung Dis 10:317-322.
- Islam MM, Tan Y, Hameed HMA, Liu Z, Chhotaray C, Liu Y, Lu Z, Cai X, Tang Y, Gao Y et al (2019) Detection of novel mutations associated with independent resistance and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect 25:1041.e1-1041.e7
- Ismail N, Rivière E, Limberis J, Huo S, Metcalfe JZ, Warren RM and Van Rie A (2021) Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis. Lancet Microbe 2:e604-e616.
- Kadura S, King N, Nakhoul M, Zhu H, Theron G, Köser CU and Farhat M (2020) Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 75:2031-2043.
- Kandler JL, Mercante AD, Dalton TL, Ezewudo MN, Cowan LS, Burns SP, Metchock B, Cegielski P, Posey JE and Global PETTS Investigators (2018) Validation of novel Mycobacterium tuberculosis isoniazid resistance mutations not detectable by common molecular tests. Antimicrob Agents Chemother 62:e00974-18.
- Keam SJ (2019) Pretomanid: First approval. Drugs 79:1797-1803.
- Khan MT, Junaid M, Mao X, Wang Y, Hussain A, Malik SI and Wei DQ (2018) Pyrazinamide resistance and mutations L19R, R140H, and E144K in Pyrazinamidase of Mycobacterium tuberculosis J Cell Biochem 120:7154-7166.
- Khoshnood S, Goudarzi M, Taki E, Darbandi A, Kouhsari E, Heidary M, Motahar M, Moradi M and Bazyar H (2021) Bedaquiline: Current status and future perspectives. J Glob Antimicrob Resist 25:48-59.
- Khosravi AD, Tabandeh MR, Shahi F and Salmanzadeh S (2021) Linezolid resistance among multidrug-resistant Mycobacterium tuberculosis clinical isolates in Iran. Acta Microbiol Immunol Hung 68:203-207.
- Kidwai S, Park CY, Mawatwal S, Tiwari P, Jung MG, Gosain TP, Kumar P, Alland D, Kumar S, Bajaj A et al (2017) Dual mechanism of action of 5-Nitro-1,10-phenanthroline against Mycobacterium tuberculosis Antimicrob Agents Chemother 61:e00969-17.
- Konno K, Feldmann FM and McDermott W (1967) Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis 95:461-469.
- Lange C, Chesov D and Heyckendorf J (2019) Clofazimine for the treatment of multidrug-resistant tuberculosis. Clin Microbiol Infect 25:128-130.
- Li D, Song Y, Zhang CL, Li X, Xia X and Zhang AM (2017) Screening mutations in drug-resistant Mycobacterium tuberculosis strains in Yunnan, China. J Infect Public Health 10:630-636.
- Li K, Yang Z, Gu J, Luo M, Deng J and Chen Y (2021) Characterization of pncA Mutations and Prediction of PZA Resistance in Mycobacterium tuberculosis Clinical Isolates From Chongqing, China. Front Microbiol 11:594171.
- Li Y, Sun F and Zhang W (2019) Bedaquiline and delamanid in the treatment of multidrug-resistant tuberculosis: Promising but challenging. Drug Dev Res 80:98-105.
- Lin W, Mandal S, Degen D, Liu Y, Ebright YW, Li S, Feng Y, Zhang Y, Mandal S, Jiang Y et al (2017) Structural basis of Mycobacterium tuberculosis transcription and transcription inhibition. Mol Cell 66:169-179.e8.
- Ma P, Luo T, Ge L, Chen Z, Wang X, Zhao R, Liao W and Bao L (2021) Compensatory effects of M. tuberculosis rpoB mutations outside the rifampicin resistance-determining region. Emerg Microbes Infect 10:743-752.
- Machado D, Coelho TS, Perdigão J, Pereira C, Couto I, Portugal I, Maschmann RDA, Ramos DF, von Groll A, Rossetti MLR et al (2017) Interplay between mutations and efflux in drug resistant clinical isolates of Mycobacterium tuberculosis Front Microbiol 8:711.
- Maningi NE, Daum LT, Rodriguez JD, Said HM, Peters RPH, Sekyere JO, Fischer GW, Chambers JP and Fouriea PB (2018) Multi- and extensively drug resistant Mycobacterium tuberculosis in South Africa: A molecular analysis of historical isolates. J Clin Microbiol 56:e01214-17.
- Marrakchi H, Lanéelle G and Quémard A (2000) InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology (Reading) 146:289-296.
- Massengo-Tiassé RP and Cronan JE (2009) Diversity in enoyl-acyl carrier protein reductases. Cell Mol Life Sci 66:1507-1517.
- McClure WR and Cech CL (1978) On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem 253:8949-8956.
- McCune RM and Tompsett R (1956) Fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. I. The persistence of drug-susceptible tubercle bacilli in the tissues despite prolonged antimicrobial therapy. J Exp Med 104:737-762.
- McNerney R, Kiepiela P, Bishop KS, Nye PM and Stoker NG (2000) Rapid screening of Mycobacterium tuberculosis for susceptibility to rifampicin and streptomycin. Int J Tuberc Lung Dis 4:69-75.
- Mikhailovich V, Lapa S, Gryadunov D, Sobolev A, Strizhkov B, Chernyh N, Skotnikova O, Irtuganova O, Moroz A, Litvinov V et al (2001) Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips. J Clin Microbiol 39:2531-2540.
- Miotto P, Cabibbe AM, Borroni E, Degano M and Cirilloa DM (2018) Role of disputed mutations in the rpoB gene in interpretation of automated liquid MGIT culture results for rifampin susceptibility testing of Mycobacterium tuberculosis J Clin Microbiol 56:e01599-17.
- Mirnejad R, Asadi A, Khoshnood S, Mirzaei H, Heidary M, Fattorini L, Ghodousi A and Darban-Sarokhalil D (2018) Clofazimine: A useful antibiotic for drug-resistant tuberculosis. Biomed Pharmacother 105:1353-1359.
- Mitchison DA (1985) The action of antituberculosis drugs in short-course chemotherapy. Tubercle 66:219-225.
- Murray JF, Schraufnagel DE and Hopewell PC (2015) Treatment of tuberculosis: A historical perspective. Ann Am Thorac Soc 12:1749-1759.
- Musser JM, Kapur V, Williams DL, Kreiswirth BN, Van Soolingen D and Van Embden JDA (1996) Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: Restricted array of mutations associated with drug resistance. J Infect Dis 173:196-202.
- Ohno H, Koga H, Kohno S, Tashiro T and Hara K (1996) Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother 40:1053-1056.
- Oliveira JS, Pereira JH, Canduri F, Rodrigues NC, de Souza ON, de Azevedo WF, Basso LA and Santos DS (2006) Crystallographic and pre-steady-state kinetics studies on binding of NADH to wild-type and isoniazid-resistant enoyl-ACP(CoA) reductase enzymes from Mycobacterium tuberculosis J Mol Biol 359:646-666.
- Pang Y, Lu J, Wang Y, Song Y, Wang S and Zhao Y (2013) Study of the Rifampin Monoresistance Mechanism in Mycobacterium tuberculosis Antimicrob Agents Chemother 57:893-900.
- Pang Y, Zong Z, Huo F, Jing W, Ma Y, Dong L, Li Y, Zhao L, Fu Y and Huang H (2017) In vitro drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing, China. Antimicrob Agents Chemother 61:e00900-17.
- Park J, Jang W, Kim M, Kim Y, Shin SY, Park K, Kim MS and Shin S (2018a) Molecular drug resistance profiles of Mycobacterium tuberculosis from sputum specimens using ion semiconductor sequencing. J Microbiol Methods 145:1-6.
- Park J, Shin SY, Kim K, Park K, Shin S and Ihm C (2018b) Determining genotypic drug resistance by ion semiconductor sequencing with the ion ampliSeqTM TB panel in multidrug-resistant Mycobacterium tuberculosis isolates. Ann Lab Med 38:316-323.
- Parveen U, Sultana S, Heba SF, Rafi R, Begum A and Fatima N (2021) Pretomanid: A novel therapeutic paradigm for treatment of drug resistant tuberculosis. Indian J Tuberc 68:106-113.
- Patel H, Pawara R, Pawara K, Ahmed F, Shirkhedkar A and Surana S (2019) A structural insight of bedaquiline for the cardiotoxicity and hepatotoxicity. Tuberculosis (Edinb) 117:79-84.
- Perrin C, Athersuch K, Elder G, Martin M and Alsalhani A (2022) Recently developed drugs for the treatment of drug-resistant tuberculosis: A research and development case study. BMJ Glob Heal 7:e007490.
- Petrella S, Gelus-Ziental N, Maudry A, Laurans C, Boudjelloul R and Sougakoff W (2011) Crystal structure of the pyrazinamidase of Mycobacterium tuberculosis: Insights into natural and acquired resistance to pyrazinamide. PLoS One 6:e15785.
- Pierattelli R, Banci L, Eady NAJ, Bodiguel J, Jones JN, Moody PCE, Raven EL, Jamart-Grégoire B and Brown KA (2004) Enzyme-catalyzed mechanism of isoniazid activation in class I and class III peroxidases. J Biol Chem 279:39000-39009.
- Plinke C, Cox HS, Kalon S, Doshetov D, Rüsch-Gerdes S and Niemann S (2009) Tuberculosis ethambutol resistance: Concordance between phenotypic and genotypic test results. Tuberculosis (Edinb) 89:448-452.
- Plinke C, Cox HS, Zarkua N, Karimovich HA, Braker K, Diel R, Rüsch-Gerdes S, Feuerriegel S and Niemann S (2010) embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother 65:1359-1367.
- Ramaswamy S and Musser JM (1998) Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis 79:3-29.
- Rawat R, Whitty A and Tonge PJ (2003) The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: Adduct affinity and drug resistance. Proc Natl Acad Sci U S A 100:13881-13886.
- Rifat D, Li SY, Ioerger T, Shah K, Lanoix JP, Lee J, Bashiri G, Sacchettini J and Nuermberger E (2021) Mutations in fbiD (Rv2983) as a novel determinant of resistance to pretomanid and delamanid in Mycobacterium tuberculosis Antimicrob Agents Chemother 65:e01948-20.
- Safi H, Sayers B, Hazbón MH and Alland D (2008) Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob Agents Chemother 52:2027-2034.
- Saint-Joanis B, Souchon H, Wilming M, Johnsson K, Alzari PM and Cole ST (1999) Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis Biochem J 338:753-760.
- Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, Morris SL and Jacobs WR (2002) A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med 8:1171-1174.
- Schrödinger LLC (2015) The {PyMOL} Molecular Graphics System. Version 1.8.
- Scorpio A and Zhang Y (1996) Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med 2:662-667.
- Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M and Zhang Y (1997) Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis Antimicrob Agents Chemother 41:540-543.
- Seifert M, Catanzaro D, Catanzaro A and Rodwell TC (2015) Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: A systematic review. PLoS One 10:e0119628.
- Selikoff IJ, Robitzek EH and Ornstein GG (1952) Treatment of pulmonary tuberculosis with hydrazide derivatives of isonicotinic acid. J Am Med Assoc 150:973-980.
- Shea J, Halse TA, Kohlerschmidt D, Lapierre P, Modestil AHA, Kearns CH, Dworkin FF, Rakeman JL, Escuyer V and Musser KA (2021) Low-level rifampin resistance and rpoB mutations in Mycobacterium tuberculosis: An analysis of whole-genome sequencing and drug susceptibility test data in new york. J Clin Microbiol 59:e01885-20.
- Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, Wang H, Zhang W and Zhang Y (2011) Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis: A potential mechanism for shortening the duration of tuberculosis chemotherapy. Science 333:1630-1632.
- Shoeb HA, Bowman BU, Ottolenghi AC and Merola AJ (1985) Evidence for the generation of active oxygen by isoniazid treatment of extracts of Mycobacterium tuberculosis H37Ra. Antimicrob Agents Chemother 27:404-407.
- Sinha P, Srivastava GN, Tripathi R, Mishra MN and Anupurba S (2020) Detection of mutations in the rpoB gene of rifampicin-resistant Mycobacterium tuberculosis strains inhibiting wild type probe hybridization in the MTBDR plus assay by DNA sequencing directly from clinical specimens. BMC Microbiol 20:284.
- Smith T, Wolff KA and Nguyen L (2012) Molecular biology of drug resistance in Mycobacterium tuberculosis Curr Top Microbiol Immunol 374:53-80
- Somoskovi A, Parsons LM and Salfinger M (2001) The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis Respir Res 2:164-168.
- Sotgiu G, Centis R, D’Ambrosio L and Battista Migliori G (2015) Tuberculosis treatment and drug regimens. Cold Spring Harb Perspect Med 5:a017822.
- Sreevatsan S, Stockbauer KE, Pan X, Kreiswirth BN, Moghazeh SL, Jacobs WR, Telenti A and Musser JM (1997) Ethambutol resistance in Mycobacterium tuberculosis: Critical role of embB mutations. Antimicrob Agents Chemother 41:1677-1681.
- Stancil SL, Mirzayev F and Abdel-Rahman SM (2021) Profiling pretomanid as a therapeutic option for TB infection: Evidence to date. Drug Des Devel Ther 15:2815-2830.
- Sun Q, Xiao TY, Liu HC, Zhao XQ, Liu ZG, Li YN, Zeng H, Zhao LL and Wan KL (2018) Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17.
- Sun Q, Li X, Perez LM, Shi W, Zhang Y and Sacchettini JC (2020) The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD. Nat Commun 11:339.
- Takawira FT, Mandishora RSD, Dhlamini Z, Munemo E and Stray-Pedersen B (2017) Mutations in rpoB and katG genes of multidrug resistant Mycobacterium tuberculosis undetectable using genotyping diagnostic methods. Pan Afr Med J 27:145.
- Takayama K and Kilburn JO (1989) Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis Antimicrob Agents Chemother 33:1493-1499.
- Tan YZ, Rodrigues J, Keener JE, Zheng RB, Brunton R, Kloss B, Giacometti SI, Rosário AL, Zhang L, Niederweis M et al (2020) Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis Nat Commun 11:3396.
- Telenti A, Philipp WJ, Sreevatsan S, Bernasconi C, Stockbauer KE, Wieles B, Musser JM and Jacobs WR (1997) The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med 3:567-570.
- Thomas JP, Baughn CO, Wilkinson RG and She I (1961) A new synthetic compound with antituberculous activity in mice: Ethambutol (dextro-2,2’-(ethylenediimino)-di-l-butanol). Am Rev Respir Dis 83:891-893.
- Thwe EP, Namwat W, Pinlaor P, Rueangsak K and Sangka A (2021) Novel mutations detected from drug resistant Mycobacterium tuberculosis isolated from North East of Thailand. World J Microbiol Biotechnol 37:194.
- Timmins GS, Master S, Rusnak F and Deretic V (2004) Nitric oxide generated from isoniazid activation by KatG: Source of nitric oxide and activity against Mycobacterium tuberculosis Antimicrob Agents Chemother 48:3006-3009.
- Unissa AN, Subbian S, Hanna LE and Selvakumar N (2016) Overview on mechanisms of isoniazid action and resistance in Mycobacterium tuberculosis Infect Genet Evol 45:474-492.
- Ushtanit A, Mikhailova Y, Lyubimova A, Makarova M, Safonova S, Filippov A, Borisov S and Zimenkov D (2021) Genetic profile of linezolid-resistant M. Tuberculosis clinical strains from moscow. Antibiotics (Basel) 10:1243.
- Vilchèze C and Jacobs WR (2007) The mechanism of isoniazid killing: Clarity through the scope of genetics. Annu Rev Microbiol 61:35-50.
- Vilchèze C and Jacobs WRJ (2014) Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: Genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013.
- Vilchèze C, Wang F, Arai M, Hazbón MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC and Jacobs WR (2006) Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med 12:1027-1029.
- Vögeli B, Rosenthal RG, Stoffel GMM, Wagner T, Kiefer P, Cortina NS, Shima S and Erb TJ (2018) InhA, the enoyl-thioester reductase from Mycobacterium tuberculosis forms a covalent adduct during catalysis. J Biol Chem 293:17200-17207.
- Wang L, Yang J, Chen L, Wang W, Yu F and Xiong H (2022) Whole-genome sequencing of Mycobacterium tuberculosis for prediction of drug resistance. Epidemiol Infect 150:e22.
- Wei CJ, Lei B, Musser JM and Tu SC (2003) Isoniazid activation defects in recombinant Mycobacterium tuberculosis catalase-peroxidase (KatG) mutants evident in InhA inhibitor production. Antimicrob Agents Chemother 47:670-675.
- Wengenack NL, Lane BD, Hill PJ, Uhl JR, Lukat-Rodgers GS, Hall L, Roberts GD, Cockerill FR, Brennan PJ, Rodgers KR et al (2004) Purification and characterization of Mycobacterium tuberculosis KatG, KatG(S315T), and Mycobacterium bovis KatG(R463L). Protein Expr Purif 36:232-243.
- Whitney JB and Wainberg MA (2002) Isoniazid, the frontline of resistance in Mycobacterium tuberculosis McGill J Med 6:114-123.
- Wilming M and Johnsson K (1999) Spontaneous formation of the bioactive form of the tuberculosis drug isoniazid. Angew Chemie Int Ed Engl 38:2588-2590.
- Wolff KA and Nguyen L (2012) Strategies for potentiation of ethionamide and folate antagonists against Mycobacterium tuberculosis Expert Rev Anti Infect Ther 10:971-981.
- Wolucka BA, McNeil MR, De Hoffmann E, Chojnacki T and Brennan PJ (1994) Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J Biol Chem 269:23328-23335.
- World Health Organization (2021) Global Tuberculosis Report 2021.
- Yang JS, Kim KJ, Choi H and Lee SH (2018) Delamanid, bedaquiline, and linezolid minimum inhibitory concentration distributions and resistance-related gene mutations in multidrug-resistant and extensively drug-resistant tuberculosis in Korea. Ann Lab Med 38:563-568.
- Yu S, Girotto S, Lee C and Magliozzo RS (2003) Reduced affinity for isoniazid in the S315T mutant of Mycobacterium tuberculosis KatG is a key factor in antibiotic resistance. J Biol Chem 278:14769-14775.
- Zhang L, Zhao Y, Gao Y, Wu L, Gao R, Zhang Q, Wang Y, Wu C, Wu F, Gurcha SS et al (2020) Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368:1211-1219.
- Zhang S, Chen J, Shi W, Liu W, Zhang W and Zhang Y (2013) Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis Emerg Microbes Infect 2:e34.
- Zhang Y and Yew WW (2015) Mechanisms of drug resistance in Mycobacterium tuberculosis: Update 2015. Int J Tuberc Lung Dis 19:1276-1289.
- Zhang Y, Heym B, Allen B, Young D and Cole S (1992) The catalase - Peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis Nature 358:591-593.
- Zhang Y, Scorpio A, Nikaido H and Sun Z (1999) Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Bacteriol 181:2044-2049.
- Zhang Y, Permar S and Sun Z (2002) Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol 51:42-49.
- Zhang Y, Wade MM, Scorpio A, Zhang H and Sun Z (2003) Mode of action of pyrazinamide: Disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother 52:790-795.
- Zhang Y, Shi W, Zhang W and Mitchison D (2014) Mechanisms of Pyrazinamide Action and Resistance. Microbiol Spectr. 2:MGM2-0023-2013.
- Zhao X, Yu H, Yu S, Wang F, Sacchettini JC and Magliozzo RS (2006) Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry 45:4131-4140.
- Zhu C, Liu Y, Hu L, Yang M and He ZG (2018) Molecular mechanism of the synergistic activity of ethambutol and isoniazid against Mycobacterium tuberculosis J Biol Chem 293:16741-16750.
- Zhu L, Bi H, Ma J, Hu Z, Zhang W, Cronan JE and Wang H (2013) The two functional enoyl-acyl carrier protein reductases of Enterococcus faecalis do not mediate triclosan resistance. mBio 4:e00613-13.
- Zloh M, Gupta M, Parish T and Brucoli F (2021) Novel C-3-(N-alkyl-aryl)-aminomethyl rifamycin SV derivatives exhibit activity against rifampicin-resistant Mycobacterium tuberculosis RpoBS522L strain and display a different binding mode at the RNAP β-subunit site compared to rifampicin. Eur J Med Chem 225:113734.
Supplementary material
The following online material is available for this article:
Table S1 - Novel KatG mutations.
Table S2 - Novel RpoB mutations.
Edited by
Associate Editor:
Publication Dates
-
Publication in this collection
23 Jan 2023 -
Date of issue
2023
History
-
Received
25 Aug 2022 -
Accepted
18 Dec 2022