Abstract
Avian cellulitis is an inflammatory process in the subcutaneous tissue, mainly located in the abdomen and thighs. This problem is commonly observed in poultry at slaughter and it is considered one of the major causes of condemnation of carcasses in Brazil. The aim of this study was to perform the microbial isolation of lesions of avian cellulitis from a processing plant located in the State of Goiás in order to analyze antimicrobial resistance by antibiogram test and to detect resistance genes by polymerase chain reaction. A total of 25 samples of avian cellulitis lesions were analyzed, from which 30 bacterial strains were isolated. There were eleven (44%) strains of Escherichia coli, nine (36%) strains of Staphylococcus epidermidis, seven (28%) strains of Proteus mirabilis and three (12%) strains of Manheimiahaemolytica. The antibiogram test showed that all strains were resistant to at least one antimicrobial. The gene of antimicrobial resistance tetB was detected in E. coli, S. epidermidis and P. mirabilis strains, and was the most frequently observed gene. The gene of antimicrobial resistance Sul1 was detected in all bacterial species, while tetA was found in E. coli and S. epidermidis strains, SHV in E. coli strains, S. epidermidis and P. mirabilis,and cat1 in one P. mirabilis strain. The results suggest a potential public health hazard due to the ability of these microorganisms to transmit antimicrobial resistancegenes to other microorganisms present in the intestinal tract of humans and animals, which may affect clinical-medical usage of these drugs.
PCR; Antibiotic susceptibility test; poultry carcasses
Antimicrobial resistance of bacterial strains isolated from avian cellulitis
Santos MMI; Alcântara ACMII; Perecmanis SI,III; Campos AIV; Santana APIII
IAluna de pós graduação (mestrado) da Universidade de Brasília (UnB)
IIAluna de Graduação Do curso de Medicina Veterinária da Universidade de Brasília (UnB)
IIILaboratório de Microbiologia de Alimentos ASS 128/10, Faculdade de Agronomia e Medicina Veterinária, Campus Darcy Ribeiro, ICC Sul, Cx. Postal 4508, Brasília, DF, Brasil
IVMinistério da Agricultura, Pecuária e Abastecimento (MAPA), Brasil
Corresponding author Corresponding author Milena M.dos Santos Aluna de pós graduação (mestrado) da Universidade de Brasília (UnB) E-mail: milenavet@hotmail.com
ABSTRACT
Avian cellulitis is an inflammatory process in the subcutaneous tissue, mainly located in the abdomen and thighs. This problem is commonly observed in poultry at slaughter and it is considered one of the major causes of condemnation of carcasses in Brazil. The aim of this study was to perform the microbial isolation of lesions of avian cellulitis from a processing plant located in the State of Goiás in order to analyze antimicrobial resistance by antibiogram test and to detect resistance genes by polymerase chain reaction. A total of 25 samples of avian cellulitis lesions were analyzed, from which 30 bacterial strains were isolated. There were eleven (44%) strains of Escherichia coli, nine (36%) strains of Staphylococcus epidermidis, seven (28%) strains of Proteus mirabilis and three (12%) strains of Manheimiahaemolytica. The antibiogram test showed that all strains were resistant to at least one antimicrobial. The gene of antimicrobial resistance tetB was detected in E. coli, S. epidermidis and P. mirabilis strains, and was the most frequently observed gene. The gene of antimicrobial resistance Sul1 was detected in all bacterial species, while tetA was found in E. coli and S. epidermidis strains, SHV in E. coli strains, S. epidermidis and P. mirabilis,and cat1 in one P. mirabilis strain. The results suggest a potential public health hazard due to the ability of these microorganisms to transmit antimicrobial resistancegenes to other microorganisms present in the intestinal tract of humans and animals, which may affect clinical-medical usage of these drugs.
Keywords: PCR, Antibiotic susceptibility test, poultry carcasses.
INTRODUCTION
Avian cellulitis was first described in Britain by Randall et al. (1984). Characterized as an acute purulent inflammation of the subcutaneous tissue, it is routinely encountered in the abdominal region and the legs of poultry (Messier et al. 1993, Gross 1994,). Carcasses presenting cellulitis should be partially or totally condemned, as determined by Ordinance No.210 of 10/11/1998 of the Brazilian Ministry of Agriculture (Brazil, 1998), which establishes the Technical Regulations of Technological and Sanitary-Hygienic Inspection of Poultry Meat. Some authors mention that condition as one of the main causes of carcass condemnation in broiler processing plants (Brito et al., 2002; Armendaris, 2006; Santana et al., 2008). Considering the importance of this pathology in the context of poultry meat production, this study aimed at performing microbiological isolation, testing antimicrobial resistance, and detecting antimicrobial resistance genes in microorganisms isolated from avian cellulitis lesions obtained from chicken carcasses stored in the refrigerator of a processing plant located in the state of Goiás, Brazil.
MATERIAL AND METHODS
1) Collection, isolation, and identification of avian cellulitis lesions
Samples of avian cellulitis lesions were collected from 25 carcasses that had been partially or totally condemned by officials of the Federal Inspection Service (SIF) of the Ministry of Agriculture and stored in the refrigerator of a processing plant located in the state of Goiás, Brazil. The carcasses presented irregular skin lesions, thickening, and color change, and were considered by SIF officials to be examples of avian cellulitis, according to Ordinance No. 210 of 11/10/1998 of the Ministry of Agriculture.
The methodology used for microbiological isolation was described by Konemman et al. (2001). Samples were duly identified and individually plated on blood agar culture medium (BioRad®) and kept at 37ºC in bacteriological incubators (Quimis®) for 24 hours. Subsequently, colonies were separated and individually plated on nutrient agar (Acumedia®) for biochemical identification. The following culture media and biochemical tests were used: oxidase test, catalase by the method of analysis and GRAM KOH test 3%, TSI (Triple Sugar Iron), urea, phenylalanine, citrate, indole, methyl red, arginine, lysine, gelatin, mannitol, trehalose, sucrose, and glucose. Gram-positive colonies were cultured in Baird-Parker culture medium (HIMEDIA®). Reading and interpretation of biochemical tests were performed according to Oliveira (2000) and Baron et al. (1994) for species identification.
2) Antimicrobial susceptibility testing and DNA extraction of isolated strains
Each isolated and identified colony was submitted to an antibiogram test carried out by the disk diffusion method, as recommended by the National Committee for Clinical Laboratory Standards (NCCLS) 2003.The tested pharmacological principles were ampicillin (AMP) 10 mg, cefazolin (CFZ) 30 mg, gentamicin (GEN) 10 mg, spiramycin (SP) 100 mg, doxycycline (DOX) 30 mg, cephalexin (CFX) 30 mg, sulfonamide (SOUTH) 200 mg, cephalothin(CEF) 30 mg, penicillin (PEN) 6 mg, enrofloxacin (ENRO) 5 mg, tetracycline (TET) 30 mg, neomycin (NEO) 30 IU, norfloxacin (NOR) 10 mg, erythromycin (ERI) 15 mg, amoxicillin (AX) and 25 mg chloramphenicol (CLO) 30 mg. The disks were purchased fromBIO-RAD®.
Strains submitted to total bacterial DNA extraction were individually plated on nutrient agar (Acumedia ®) and incubated in a bacteriological incubator at 37ºC for 24 hours. Subsequently, three colonies were selected and inoculated into 15 mL falcon tubes containing 3 mL of L-broth (1% peptone, casein, yeast extract, and 0.5% sodium chloride 1%) and were maintained in a shaker (New Brunswick Scientific Edison, NJ, USA) at 200 rpm for 12 hours at 37ºC. One mL of the L-broth was used for total DNA extraction, applying the phenol-chloroform (1:1) method, according to the protocol described by Sambrook et al. (2001). Total DNA was read in agarose gel at 0.8% with the addition of ethidium bromide at a concentration of 5mg/mL under ultraviolet light (Majorscience®).
3) Detection of antibiotic resistance genes
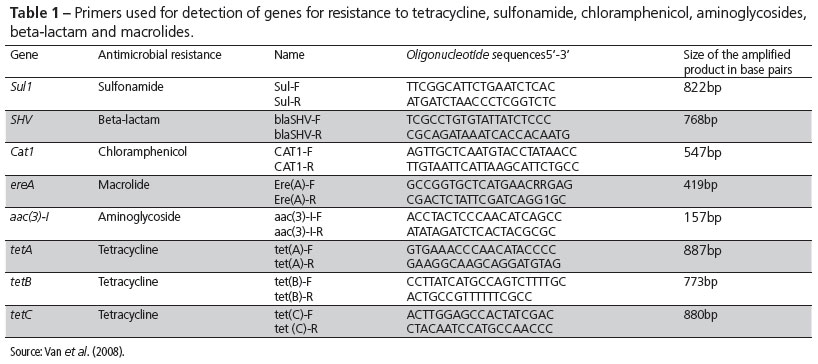
The isolated bacterial colonies were individually used to perform the PCR-based protocol described by Van et al. (2008), with minor modifications, in which simple PCR reactions were developed to detect individual tetracycline resistance genes (tetA, tetB and tetC), aminoglycoside resistance gene (aac (3 ')-I), and macrolide resistance gene (ereA). A single reaction was developed for simultaneous detection (multiplex PCR) of genes of resistance to sulfonamides (Sul1), beta-lactams (VHS), and chloramphenicol (cat1). The oligonucleotide probes used for each resistance gene are listed in Table 1.
The PCR for the detection of single tetracycline resistance genes tetA, tetB and tetC involved a total volume of 25 µl reaction containing 10ng of DNA extracted from each bacterial strain isolated from cellulitis samples, 10 pmol of each forward and reverse primer (RTD®), final concentration 1.5 mM MgCl2, final concentration 2.0 mMdNTPs (Invitrogen®) and 1 U Taq (Invitrogen®). The amplification conditions in the thermal cycler (BioRad®) were as follows: initial denaturation at 94ºC for 5 minutes, 30 cycles of denaturation at 94ºC for 30 seconds, annealing at 50ºC for 30 seconds, extension at 72ºC for 1 minute, and a final cycle of amplification at 72ºC for 10 minutes. The protocol used to detect the macrolide resistance gene (ereA) and the aminoglycoside resistance gene (aac (3')-I) was the same as that described for the tetracycline resistance gene, except forMgCl2 final concentration, which was 2.5 mM, and annealing temperature, which was 59ºC for the gene ereA and 55ºC for the gene aac(3 ')-I. In the original protocol of Van et al. (2008), the PCR of these two resistance genes (ereA and aac (3')-I) was performed in a multiple reaction with other resistance genes.
The PCR for the simultaneous detection (multiplex PCR) of the sulfonamide, chloramphenicol and beta-lactams resistance genes (sul1, cat1 and SHV) was performed in a total volume of 25 µl reaction containing 10ng of DNA extracted from each isolated strain, with final concentration of 3.0 mM MgCl2. This concentration was modified, as in the original protocol proposed by Van et al. (2008), the concentration was 4.0 mM. A final concentration of 2.0 mMdNTP was used, with 10 pmol of each reverse and forward primer and 1 U of Taq polymerase (Invitrogen®). Amplification conditions were denaturation at 94ºC for 15 minutes, followed by 30 cycles of denaturation at 94ºC for 30 seconds, annealing at 59ºC for 30 seconds - the annealing temperature was 58ºC in the original protocol described by Van et al. (2008) - and extension at 72ºC for 1 minute and one final cycle of amplification at 72ºC for 10 minutes. PCR products were visualized and photo documented (Majorscience®).
RESULTS
1) Microbiological isolation from samples of avian cellulitis
Out of the 25 analyzed samples of avian cellulitis, 11 (37%) strains of Escherichia coli, 9 (30%) of Staphylococcus epidermidis, seven (23%) of Proteus mirabilis and 3 (10%) of Manheimia haemolytica were isolated. A total of 30 bacterial strains, and two bacterial genera were isolated from some samples of cellulitis. The results of bacterial isolation in this study revealed the presence of more than one type of microorganism present in the lesions of avian cellulitis obtained from the processing plant.
2) Detection of resistance genes in isolates from avian cellulitis lesions
The results of antimicrobial resistance genes are shown in Table 2 and Figure 1. One strain of E. coli and one of P. mirabilis that showed the gene for beta-lactam resistance (SHV) were also phenotypically resistant to this class of antimicrobial agents (amoxicillin, ampicillin and penicillin). One strain of E. coli and two strains of S. epidermidis were positive for the gene Sul1, and these were phenotypically sensitive to the drug. Two of the seven P. mirabilis strains were resistant to sulfonamide. The P. mirabilis strain that was phenotypically resistant to chloramphenicol presented the resistance gene to this antimicrobial agent (cat1). Macrolide-resistant genes (ereA) and those resistant to aminoglycosides (aac (3) -1) were not detected in any of the isolated bacterial species.
DISCUSSION
The results of bacterial isolation in this study revealed the presence of more than one type of microorganism present in the samples of avian cellulitis lesions collected from carcasses stored in a refrigerator of a processing plant located in the state of Goiás. This is in contrast with the study by Vieira et al. (2005), who reported the isolation of only Escherichia coli in all 20 (100%) samples of avian cellulitis lesions collected from broiler carcasses stored in a refrigerator of a processing plant in the state of Rio de Janeiro. Andrade (2005) also found Escherichia coli in cellulitis lesions of broiler carcasses in the state of São Paulo. In a study conducted in England with broilers, Randall et al. (1984) were the first to report an association between Escherichia coli and Pasteurella multocida in avian cellulitis lesions. Messier et al. (1993) also isolated Escherichia coli and Streptococcus dysgalactiae in cellulitis samples of chicken carcasses in Canada. In a study on cellulitis in chicken carcasses in Iran, Derakhshanfar et al. (2002) isolated Escherichia coli in 91.8% of the carcasses, and reported that E. coli strains were associated or not with Staphylococcus aureus. Actinomyces pyogenes and Staphylococcus aureus were also isolated as the sole agent in some samples. In general, research on avian cellulitis shows that Escherichia coli is the most frequently encountered bacterium in this type of lesion.
The results of the present study are consistent with the findings of other authors relative to the phenotypic resistance to antibiotics. Working with E. Coli strains isolated from airsacculitis, pericarditis and tracheitis lesions sampled from broilers in the state of Rio de Janeiro, Gonçalves et al. (2005) found 100% resistance to penicillin, erythromycin, and other antimicrobial agents. In his review of studies on the antimicrobial resistance profile of E. coli strains in chickens conducted in the United States, Canada, France, Australia and Iceland, Gyles (2008) mentioned that Avian Pathogenic E. coli (APEC) strains were often found to be highly resistant to tetracycline, sulfonamide, and streptomycin. Xin-Sheng et al. (2007), in a study of 70 strains of E. coli isolated from the livers of chickens reared on different farms in China, reported high resistance to ampicillin (83%). Zanatta et al. (2004) found similar results in a study with 120 E. coli samples obtained in the necropsy of commercial broilers in Descalvado, SP, Brazil, out of which 54.6% were resistant to cephalexin and cephalothin.
Resistance to chloramphenicol was detected in 18.19% of the isolated E. coli strains, which was lower compared with the other tested antimicrobial agents tested in this study. This may explained by the fact that the use of chloramphenicol in animal production is forbidden since 2003 in Brazil(Brazil, 2003).Brito et al. (2000), in a study of 10 E. coli strains isolated from cellulitis lesions in quails reared in commercial farms in northern region of the state of Paraná, observed that 90% of the isolated strains were resistant to chloramphenicol.
To the best of our knowledge, there are no reports of microbiological isolation of Proteus mirabilis, Staphylococcus epidermidis and Manheimia haemolytica from avian cellulitis lesions. However, some authors have reported the occurrence of antimicrobial resistance of these organisms isolated from other sources. Shin-hee et al. (2005), in a study with ground beef, turkey, chicken and pork, isolated 64 multi-resistant strains of Proteus mirabilis from a commercial establishment located in Stillwater, Oklahoma, United States, but only from samples of homogenized turkey, chicken and pork meat. The isolated strains were resistant to at least four antimicrobial agents, especially to ampicillin, tetracycline, gentamicin and kanamycin. Qiongfen et al. (2011), in a study of stool samples from pigs reared on a farm located in eastern China, found a single P. mirabilis strain that was resistant to chloramphenicol, trimethoprim/sulfamethoxazole and tetracycline. In a study conducted in France, Even et al. (2010) isolated 33 strains of Staphylococcus epidermidis from cheese and dry fermented sausages samples, and clinical samples from hospitalized patients, and reported that 73% were resistant to at least one of the antimicrobials: 9.18% to tetracycline, 36% to erythromycin, 39% to penicillin, and 18% to norfloxacin, among other antimicrobial agents. Onni et al. (2011), in a study of 131 strains of S. epidermidis isolated from mastitis in goats reared in different geographical areas of the region of Sardinia, Italy, found that 38% of the strains were resistant to penicillin, 7.6% were resistant to tetracycline, and 2.3% resistant to both penicillin and tetracycline.
Some authors isolated Manheimia haemolytica strains from sources other than avian cellulitis. Klima et al. (2011), in a study with bovine nasopharyngeal samples collected at the beginning and end of feedlot in two farms in southern Alberta, Canada, isolated of M. haemolytica strains and found that all isolates were resistant to sulfametazole/trimethoprim, ceftiofur, enrofloxacin, florfenicol and gentamicin. In that study, the authors demonstrated that out of the 409 Manheimia haemolytica isolates, 39 (9.54%) were resistant to only one type of antimicrobial agent, and five (1.2%) were resistant to more than one antimicrobial agent. Hendriscken et al. (2008) conducted a survey of data on the antimicrobial susceptibility of pathogenic bacteria, including M. haemolytica isolated from samples of cattle reared in different European countries between 2002 and 2004.
Further studies should be conducted to determine true origin of the antimicrobial resistance observed in present study. The resistance of bacteria isolated from this type of lesion may pose a considerable public health hazard, because these lesions may be a source of cross-contamination of chicken carcasses during processing, disseminating antimicrobial-resistant bacteria, when the carcasses are only partially used.
Relative to the results obtained in the detection of antimicrobial resistance genes, in a study of E. coli isolated strains from broiler chickens in Vietnam, Van et al. (2008) observed the concomitant presence of tetA and tetC, and tetB and tetC genes. This is consistent with the detection of two concomitant resistance genes in the same strain of E. coli in the present study. Xin-Sheng et al. (2007) detected the presence of the cat1 gene in 11 of 70 E. coli strains isolated from livers of sick chickens reared in China. Abdullah et al. (2010), in a study with samples of raw chicken meat sold in supermarkets of the city of Taif, in Saudi Arabia, detected the presence of the Sul1 gene in all isolated E. coli strains. Soufi et al. (2009) also detected sulfonamide (Sul1) and cetyltransferase (CAT) resistance genes in of E. coli strains isolated from chicken and turkey meat sampled in a processing plant in Tunisia. Simeoni et al. (2008), in a study with samples obtained at various processing stages in two pig processing plants of northern Italy, isolated S. epidermidis, which presented tetracycline (tetM, tetO, and tetK), beta-lactams (blaZ), aminoglycosides (aac (6 ') aph2), methicillin (mecA), and macrolidelincosamide-streptogramins (ermA, ermB and ermC) resistance genes. The presence of multidrug-resistant bacteria was also observed in the present study.
There was general consistency between the results obtained in the analysis of phenotypic antimicrobial resistance and the detection of resistance genes. The presence of the chloramphenicol resistance gene may suggest that this gene is still present in bacterial strains, although its use is forbidden in Brazil since 2003. According to Even et al. (2010), S. epidermidis in foods may be a significant reservoir of antimicrobial resistance genes, and may be involved in resistance transfer among microorganisms present in foodstuffs. Further studies should be conducted to establish the origin and the possibility resistance transfer by microorganisms in foodstuffs.
CONCLUSION
The present study identified Escherichia coli, Staphylococcus epidermidis, Proteus mirabilis, and Manheimiahaemolyica strains in samples of avian cellulitis lesions. The presence of genes for resistance to antimicrobial agents was detected by PCR in those four bacterial species. This is the first study that isolated and detected resistance genes to antimicrobial agents in this type of lesion in Brazil. The presence of antimicrobial resistance in bacteria isolated from a feedstuff intended for human consumption was demonstrated. Further studies are needed to verify the origin of this resistance and to analyze the probable risk carcass contamination at the time of inspection when only part of the carcass is used.
ACKNOWLEDGEMENTS
The authors are grateful to the agency Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the M. Sc. Scholarship given to the first author and to Decanato de Pesquisa e Pós-graduação (DPP) of the University of Brasilia (UNB) for the financial support.
Submitted: February/2013
Approved: July/2013
- Abdullah DA, Youssuf AG, Sabry AH.Antibiotic Resistance in Escherichia coli Isolated from Retail Raw Chicken Meat in Taif, Saudi Arabia.Foodborne Pathogens and Disease 2010; 7(3):281-285.
- Armendaris P. Poultry Slaughter: Data sentencing, federal inspection service. In: Proceedings of the 5th Symposium of Poultry Health UFS 5.; 2006; Santa Maria, Rio Grande do Sul. Brasil. p.69-81.
- Andrade LC. Histopathology and identification of Escherichia coli as a causative agent of avian cellulitis in broiler chickens.Niteroi: Fluminense Federal University; 2005.
- Baron, E. J. Bailey and scott's diagnostic. Microbiology. St. Louis: Mosby; 1994.
- Brazil. Portaria Nº 210 de 10 de novembro de 1998. Provides for Technical Regulation of Inspection Technology and Hygiene and Sanitation Poultry. Brasília: Ministry of Agriculture and Supply.
- Brito BG, Tamehiro YC, Okano W, Luzardo MM, Berbel MM, Guimarães GI. Cervical cellulitis in broilers chickens for Escherichia coli.Semina: Agricultural Sciences 2002; 23(1):81-84.
- Derakhshanfar A, Ghanbarpour RA. Study on avian cellulitis in broiler chickens.Veterinarskiarhiv 2002;72(5):277-284.
- Even S, Leroy S, Charlier C, Zakour BN, Chacornac PJ, Lebert I, Jamet E, Desmonts HM, Coton E, Pochet S, Donnio PY, Gautier M,Talon R, Loir LY. Low occurrence of safety hazards in coagulase negative staphylococci isolated from fermented foodstuffs. International Journal of Food Microbiology 2010;139:87-95.
- Goncalves PMR. Escherichia coli iss gene detection by PCR, mycoplasma and salmonella on the sanitary quality of broilers at slaughter [Thesis]. Niteroí (RJ): Universidade Federal Fluminense; 2005.
- Gross WG. Diseases due to Escherichia coli in poultry. In: Gyles CL. Escherichia coli in domestic animals and humans. Oxon: CAB International; 1994. p.237-259.
- Gyles LC. Antimicrobial resistance in selected bacteriafrom poultry. Animal Health Research Reviews 2008;9(2):149-158.
- Hendriksen RS, Mevius D, Schroeter A, Teale C, Meunier D, Butaye P, Franco A, Utinane A, Amado A, Moreno M, Greko C, Stärk K, Berghold C, Myllyniemi LA, Wasyl D, SundeM, Aarestrup MF. Prevalence of antimicrobial resistance among bacterial pathogensisolated from cattle in different European countries: 2002-2004. Acta Veterinaria Scandinavica 2008;50:28.
- Klima CL, Alexander TW, Read RR, Gow SP, Booker CW, Hannon S , Sheedy C, McAllister TA, Selinger LB. Genetic characterization and antimicrobial susceptibility of Mannheimiahaemolytica isolated from the nasopharynx of feedlot cattle. Veterinary Microbiology 2011;149:390-398.
- Konemman EW, Allen SD, Janda WM, Scherckenberger PC, Winn JWC. Microbiological diagnosis. 5th ed. Orlando: Medicini; 1465p.
- Messier S, Quessy S, Robinson Y, Devriese LA, Hommez J, Fairbrother JM. Focal dermatitis and cellulitis in broiler chickens: bacteriological and pathological findings. Avian Disease 1993;37(3):839-44.
- National Committee for Clinical Laboratory Standards. Wayne; 2003.
- Oliveira SJ. Veterinary microbiology. Bacteriological practical g uide.2nd ed.. Canoas: Ed ULBRA;2000.
- Onni T, Sanna G, Larsen J, Tola S. Antimicrobial susceptibilities and population structure of Staphylococcus epidermidis associated with ovine mastitis. Veterinary Microbiology 2011 2011;148(1):45-50.
- Peighambari SM, Vaillancourt PJ, Wilson AR, Gyles CL. Escherichia coli cellulitis: experimental infections in broiler chickens. Avian Disease 1995;39:125-134.
- Qiongfen Y, Zhenling Z, Jianxia H, Yuting D, Liangying H, Wei T, Hongqing Z, Zhangliu C, Jian-Hua L* . Dissemination of the gene rMTBCarried on IncF and INCN plasmids among Enterobacteriaceae in a pig farm and its environment. Journal of Antimicrobial Chemotherapy 2011; 66:2475-247.
- Randall CJ, Meakins PA, Harris MP, Watt DJ. A new skin disease in broilers? Veterinary Record 1984;10;114(10):246.
- Sambrook J, Russel WD. Condensed from molecular cloning protocols. New York: Cold Spring Lab Press; 2001.
- Santana AP. Murata SL, Guimarães FC, Delphino KM, Pimentel MC. Causes of condemnation of carcasses from poultry in slaughterhouses located in State of Goias, Brazil. Rural Science, 2008;38(9).
- Simeoni D, Rizzotti L, Cocconcelli P, Gazzola S, Dellaglio F, Torriani S. Antibiotic resistance genes and identification of Staphylococci collected from the production chain of swine meat commodities. Food Microbiology 2008;25:196-201.
- Shin-hee K Cheng-i W, Haejung A. Multidrug-Resistant Klebsiella pneumoniae Isolated from Farm Environments and Retail Products in Oklahoma. Journal of Food Protection 2005;68 (10):2022-2029.
- Soufi L, Abbassi MS, Sáenz Y, Vinué L, Somalo S, Zarazaga M, Abbas A, Dbaya R, Khanfir L, Ben Hassen A, Hammami S, Torres C. Prevalence and Diversity of integrons and Associated Resistance Genes in Escherichia coli isolates from Poultry Meat. Foodborne Pathogens in Tunisia and Disease 2009;6(9):1067-73.
- Van TT, Chin J, Chapman T, Tran LT, Coloe PJ. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. International Journal of Food Microbiology 2008;10;124(3):217-23.
- Vieira TB, Franco MR, Magalhães H, Praxedes SIC, Tortelly R.Cellulitis in broiler chickens slaughtered under sanitary inspection: pathological aspects associated with the isolation of Escherichia coli. Re.Bras.Ci.Vet., set/dez. 2005 v.13 n.3, p.174-177.
- Xin-sheng L,Gui-Qin W, Xiang-Dang D, Bao-An C, Su-Mei Z, Jian-Zhong S. Antimicrobial susceptibility and molecular detection of chloramphenicoland florfenicol resistance among Escherichia coli isolates from diseased chickens. Journal Veterinary Science 2007;8(3): 243-247.
- Zanatta GF, Kanashiro AMI, Castro AGM , Cardoso ALSP., Tessari ENC, Pulic SCP. Susceptibility of Escherichia coli strains of avian origin to antimicrobials. Arquivo do Instituto Biológico 2004;71(3):283-286.
Corresponding author
Publication Dates
-
Publication in this collection
16 May 2014 -
Date of issue
Mar 2014
History
-
Received
Feb 2013 -
Accepted
July 2013