ABSTRACT
Polymorphism of three quail communities was analyzed by using 12 microsatellite markers in this paper, aiming to provide scientific references for the evaluation, protection and utilization of quail genetic resources in China. Results demonstrated that the number of observed alleles by 12 microsatellite markers ranges between 4~7. The average polymorphism information contents (PIC) of the Chinese yellow quail, the Chinese black quail and the Korean quail, as detected by 12 microsatellite markers, are 0.6853, 0.6401 and 0.6565,respectively, and average heterozygosity values are 0.7333, 0.6957 and 0.7111, respectively. This indicates that the Chinese yellow quail has the richest genetic polymorphism. According to cluster analysis, the Chinese black quail and the Korean quail have the smallest genetic distance (0.0628), which reflects that they have the closest genetic relationship. The genetic distance between the Chinese yellow quail and the Korean quail is 0.0951. Therefore, the Chinese black quail and the Korean quail are clustered together firstly, and then the Chinese yellow quail.
Keywords:
Chinese black quail; Microsatellite marker; Polymorphism; Genetic diversity
INTRODUCTION
Microsatellite markers are good genetic markers for studying genetic variation in a same species and among different species. Due to their large quantity, wide and uniform distribution, rich polymorphism, codominant inheritance, as well as simple and convenient analysis method, microsatellite markers are molecular markers that are widely used to evaluate genetic resources of livestock at present.
Microsatellite markers are mainly used in quail breeding for the establishment of the quail genetic map. Kayang et al. (2004Kayang BB, Vignal A, Inoue-Murayama M, Miwa M, Monvoisin JL, Ito S,Minvielle F. A first-generation microsatellite linkage map of the Japanese quail. Animal Genetics 2004 ;35: 195-200.) developed the first generation of microsatellite linkage maps with 72 microsatellite markers based on 100 microsatellite markers discovered in quail genomes. Quail functional gene mapping and QTL: in 2005, Miwa et al. located three blood protein loci Tf, Hb-1 and Pa-1 on QL08, CJA14 and QL13 chromosomes by using three microsatellite primers GUJ0071, GUJ0097 and GUJ0061 (Miwa et al., 2005).Genetic diversity analysis of quails: many polymorphic analyses on wild quail and domesticated quail using microsatellite markers are reported in both China and foreign countries. Studieshave accumulated data on the evaluation of quail genetic resources and analyzed population genetic variation and evolutionary relationship (Wang et al., 2004Wang YH, Chang H, Xu W, Chang GB. Genetic analysis of microsatellite DNA markers in domestic quail and wild Japanese quail populations. Vcta. Veterinariaet. Zootechnica. Sinica 2004;4:367-371.; Olowofeso et al., 2006; Chang et al., 2007Chang GB, Chang H, Liu1 XP, Zhao WM, Ji DJ, Mao YJ, Song GM, Shi XK.Genetic diversity of wild quail in China ascertained with microsatellite DNA markers.Asian-Aust. J. Anim. Sci. 2007; 20(12) : 1783 -1790.; Amirinia, 2007Amirinia.Evaluation of eight microsatellite loci polymorphism in four Japanese quail (Coturnix japonica)strain in Iran. Pakistan Journal of Biological Sciences 2007;10(8): 1195-1199.; Kim et al., 2007Kim SH, Cheng MT, Ritland K, Silvesides FG Inbreeding in Japaneses Quail estimated by Microsatellite analyses. J. Hered 2007;98:378-381.; Olympe et al., 2010Olympe C, Francis M, Denis R, Bertrand B ,Katia F,Jean-Luc C, Boniface BK, Sophie L,Alain V, Jean-Marie B, Xavier R. Evidence for introgressive hybridization of wild common quail (Coturnix coturnix) by domesticated Japanese quail (Coturnix japonica) in France. Conserv Genet 2010;11:1051-1062.; Wu et al., 2010Wu SJ.Evalution of Genetic diversity among four feathers eggs quail populations by using microsatellites.luoyang in China: Master degree thesis of Henan University of Science and Technology,2010.; Hossein et al., 2011Hossein E, Cyrus A, Mohammad AR. Genetic variation and bottleneck in japanese quail (Coturnix japonica) strains using twelve microsatellite markers[J]. African Journal of Biotechnology 2011;20: 4289-4295.; Farrag et al., 2011Farrag SA, Tanatarov AB, Soltan ME, Ismail M, Zayed OM.Microsatellite analysis of genetic diversity in three populations of japanese quail (Coturnix coturnix japonica) from kazakhstan[J]. J Anim Vet Adv 2011;18:2376-2383.; Thakur et al., 2011Thakur M,Ishwari DR, Rishi PM,Sambandam S. A panel of polymorphic microsatellite markers in Himalayan monal Lophophorus impejanus developed by cross-species amplification and their applicability in other Galliformes. Eur J Wildl Res 2011;57:983-989.; Bai et al., 2013Bai J, PangY, Wu S, Yu M, Zhang X, Zhao S, Xu H.Polymorphism analysis of Chinese yellow quail uing microsatellite markers[J]. J Anim Plant Sci 2013;23(4): 1072-1076.).
The quail is an ancient bird and is also called Japanese quail. There are mainly wild quails and domesticated quails. Wild quails can be divided into wild common quail and wild Japanese quail. Although China started quail productionlate, it achieved rapid development in 1970s, with unprecedented growth and diversified varieties. Both varieties of wild quails are present in China, but there is a larger number of Japanese quails (Chang et al.,2004). Korean quails, an egg-laying variety of the Japanese quail, can be divided into Longcheng and Huangcheng varieties. After being introduced in China, the Korean Longcheng variety originated the Chinese white-feather quail and the Chinese yellow quail. The Chinese yellow quailis a recessive yellow-feather variety, which was discovered and cultivated by Yue Genhua, a young teacher of Nanjing Agricultural University in 1990(Yue et al.,1994). The Chinese black quail is a feather color mutant recently discovered by our research group. It is the hybrid of male Chinese yellow quail and female Korean quail. The hybridization test confirmed that the Chinese black quail is the consequence of incomplete autosome recessive (Pang et al.,2013Pang YZ, Wang LS, Zhao SJ, Zhang XH, Li XS, Yu MQ. Genetic mechanism of black plumage gene in layer quail and its interaction with maroon-plumage gene. China Poultry 2013;35(15):6-10.).
Due to few available studies on the Chinese black quail, this study employed 12 microsatellite markers for polymorphic analysis of three quail communities (Korean quail, Chinese yellow quail and Chinese black quail) in order to discuss their evolution degree and provide scientific basis for the evaluation, protection, and utilization of Chinese black quail genetic resources.
MATERIALS AND METHODS
The tested quails derivedfrom the test farm of Henan University of Science and Technology. One hundred Chinese black quail mutants (50 males and 50 females), 75 Chinese yellow quails and 75 Korean quails (40 males and 35 females each) were randomly selected, totaling 250 quails.
Two mL of heart blood was collected into tubes with acid-citrate-dextrose (ACD ) anticoagulantsolution at 6:1 blood to ACD concentration. Blood sampleswere stored in refrigerator at -20 ºC until analyses. Genomic DNA was extracted with the blood tissue genomic DNA extraction kit (Tiangen, Beijing, China).
Twelve microsatellite primers were synthesized by Shanghai Sangon Biological Engineering Technology Co. Primer sequences are shown in Table 1.The total size of the PCR reaction system was 12.5μL, including 8.65μL of ddH2O, 1.25μL of 10×buffer, 0.75μL of Mg2+(25 mmol/L), 0.5μL of DNA template, 0.5μL (10 mmol/L) of upstream and downstream primers, 0.25μL of dNTPs, and 0.1μL of Taq enzyme. The PCR amplification process was as follows: denaturation for 3 min at 95°C; denaturation for 45 s at 94°C, annealing for 60s at X°C, extension for 60s at 72°C and 30 cycles, extension for 12 min at 72°C, and preserving at 4°C. The annealing temperature is shown in Table 1. The PCR products were processed with 10% native polyacrylamide gel electrophoresis for 6~8h under stable voltage 150~180V, and then fixed for silver nitrate staining, development, and other processes. Finally, in the gel imaging system under the photo shoot.
The allele frequency and size range of alleles were calculated using theExcel Microsatellite Toolkit. The molecular biology software POPGENE (Version1.32) was used to analyze polymorphism information content (PIC), effective number of alleles (Ne), and miscellaneous heterozygosity (H) of each marker.
Heterozygosity:
Where pi was the frequency of ith allele of a microsatellite DNA
Effective number of alleles:
Where pi was the frequency of ith and jth allele of a microsatellite DNA
Polymorphic information content:
Where pi andpj were the frequency of ith allele of a microsatellite DNA,and n is allele number.
Genetic differentiation coefficient:
Gst=1-Hs/Ht
Where Ht is total population average heterozygosity, Hs is average heterozygosity of different quail populations, and Gst is coefficient of gene differentiation.
RESULTS AND DISCUSSION
The test results of polymorphism of microsatellite marker UBC0006 in Chinese black quailsarepresented in Figure 1, which shows that the microsatellite marker UBC0006 in Chinese black quails has a relatively rich polymorphism, with five alleles detected.
The allele frequencies of 12 microsatellite markers in the three quail communities are listed in Table 2. The number of observed alleles ranged between 4~7. A total of 197 observed alleles were detected. The maximum number of GUJ0055 and GUJ0071 alleles observed in Chinese yellow quail and Chinese black quailwas 7, and the maximum number of GUJ0086 alleles observed in Korean quail was 7.
The number of observed alleles, number of effective alleles, PIC, and fixation indexes of 12 microsatellite markers in the three quail communities are shown in Table 2. The average number of observed alleles in Chinese yellow quails, Chinese black quails and Korean quailswere 5.6667, 5.3333 and 5.4167, respectively. Out of the 12 microsatellite markers, UBC0004 presented the least effective alleles (2.2321 in Chinese yellow quails) and GUJ0071 showed the most effective alleles (5.9862 in Chinese yellow quails). The average numbers of effective alleles in Chinese yellow quails, Chinese black quails and Korean quails were 3.9215, 3.3778, and 3.4591, respectively. Their mean PICs were 0.6853, 0.6401 and 0.6565, respectively. Chinese yellow quails had slightly higher PIC than the other two varieties.
Expected heterozygosity and average heterozygosity in the three quail communities are shown in Table 2. The average heterozygosity of 12 microsatellite markers in the quail communities were 0.7333, 0.6957 and 0.7111, indicating that all three quail communities present high polymorphism. The Chinese yellow quail showed the richest genetic polymorphism, while the Chinese black quail had lower genetic polymorphism. During the genetic research of a community, the Hardy-Weinberg equilibrium state test is necessary to determine whether there is population genetic equilibrium. Table 2 shows that,althoughGUJ0054 complied with theHardy-Weinberg law in Korean quails, the other markers deviated significantly or extremely significantly from the Hardy-Weinberg law. In other words, genes of these markers were in Hardy-Weinberg imbalance state.
Polymorphism information content (PIC) expresses possibility of an offspring to obtain the same allelic marker from its father (or mother). It is a statistical magnitude used to describe the heterogeneity degree of microsatellite locus, and it is an ideal evaluation index of polymorphism of an allele segment. When PIC>0.5, the locus is a highly polymorphic locus. When 0.25<PIC<0.5, the locus is moderately polymorphic. When PIC<0.25, the locus is a low polymorphic locus. In this study, all microsatellite markers belonged to highly polymorphic loci, except for UBC0004, which showed the smallest PIC (0.4698) and belonged to a moderately polymorphic locus. The average PICs of the 12 microsatellite markers in Chinese yellow quails, Chinese black quails, and Korean quails were 0.6853, 0.6401 and 0.6565, respectively, and all were higher than 0.5. The average PIC of Korean quaisl was similar to the findings (0.6945)of Meng et al. (2007).
Heterozygosity reflects an approximatedegree of genetic variation. Higher heterozygosity is accompanied of higher genetic diversity in the community and of higher degree of genetic variation. Heterozygosity of the evaluated communities and calculated using microsatellite markers was generally between 0.3 and 0.8. In this studies, 11 microsatellite markers reported high heterozygosity in the three evaluated quail communities, except for UBC0004. This indicates that all three quail communities present high polymorphism and that the Chinese yellow quail has the richest genetic polymorphism.
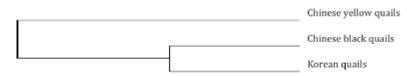
The coefficient of genetic differentiation (GST) reflects the degree of genetic variation among communities. It varies from 0~1. When communities have almost same effective alleles, no differentiation can be observed when GST approaches 0, but evident differentiation exists when GST approaches 1. Table 3 shows that the GST of the three studied quail communities varied between 0.0103~0.0773, indicating low differentiation, but high homology among them. Table 3 shows that the genetic distance between the Chinese black quail and the Korean quail was the shortest, of 0.0628. This implies that the Chinese black quail and the Korean quail have the closest genetic relationship. The second shortest genetic distance was between the Chinese yellow quail and the Korean quail, of 0.0951. Figure 2 further confirms that the Chinese black quail and the Korean quail cluster first, and then the Chinese yellow quail.
ACKNOWLEDGEMENTS
This research was supported by Henan University of Science and Technology the High Level Project Cultivation Fund Project (2015GJB028).
REFERENCES
- Amirinia.Evaluation of eight microsatellite loci polymorphism in four Japanese quail (Coturnix japonica)strain in Iran. Pakistan Journal of Biological Sciences 2007;10(8): 1195-1199.
- Bai J, PangY, Wu S, Yu M, Zhang X, Zhao S, Xu H.Polymorphism analysis of Chinese yellow quail uing microsatellite markers[J]. J Anim Plant Sci 2013;23(4): 1072-1076.
- Chang GB, Chang H, Liu XP, WangH Y, Xu W, Zhao WM,Wang QH.Study on genetic diversity of wild quail in China with microsatellite DNA markers. Acta. Genet. Sin 2005;8: 795-803.
- Chang GB, Chang H, Liu1 XP, Zhao WM, Ji DJ, Mao YJ, Song GM, Shi XK.Genetic diversity of wild quail in China ascertained with microsatellite DNA markers.Asian-Aust. J. Anim. Sci. 2007; 20(12) : 1783 -1790.
- Farrag SA, Tanatarov AB, Soltan ME, Ismail M, Zayed OM.Microsatellite analysis of genetic diversity in three populations of japanese quail (Coturnix coturnix japonica) from kazakhstan[J]. J Anim Vet Adv 2011;18:2376-2383.
- Hossein E, Cyrus A, Mohammad AR. Genetic variation and bottleneck in japanese quail (Coturnix japonica) strains using twelve microsatellite markers[J]. African Journal of Biotechnology 2011;20: 4289-4295.
- Kayang BB, Vignal A, Inoue-Murayama M, Miwa M, Monvoisin JL, Ito S,Minvielle F. A first-generation microsatellite linkage map of the Japanese quail. Animal Genetics 2004 ;35: 195-200.
- Kim SH, Cheng MT, Ritland K, Silvesides FG Inbreeding in Japaneses Quail estimated by Microsatellite analyses. J. Hered 2007;98:378-381.
- Meng QM, Sun YQ, Li DQ, Qiao AJ.Genetic diversity analysis of Korean quail using microsatellite DNA markers.Fujian J. Anim. Husb. Vet. Med2007; 1:1-2.
- Miwa M, Inoue-Murayama M, Kayang BB, Vignal A, MinvielleF, Monvoisin JL, Takahashi H, Ito S. Mapping of plumage colour and blood protein loci on the microsatellite linkage map of the Japanese quail. Animal Genetics 2005 ;36:396-400.
- Olowofeso O, Dai GJ, Wang JY, Xie KZ, Li NC, He YQ. Detection of genetic diversity of four quail populations in East China based on three microsatellite markers.J. Yangzhou University2006; 1:29-32.
- Olympe C, Francis M, Denis R, Bertrand B ,Katia F,Jean-Luc C, Boniface BK, Sophie L,Alain V, Jean-Marie B, Xavier R. Evidence for introgressive hybridization of wild common quail (Coturnix coturnix) by domesticated Japanese quail (Coturnix japonica) in France. Conserv Genet 2010;11:1051-1062.
- Pang YZ, Wang LS, Zhao SJ, Zhang XH, Li XS, Yu MQ. Genetic mechanism of black plumage gene in layer quail and its interaction with maroon-plumage gene. China Poultry 2013;35(15):6-10.
- Thakur M,Ishwari DR, Rishi PM,Sambandam S. A panel of polymorphic microsatellite markers in Himalayan monal Lophophorus impejanus developed by cross-species amplification and their applicability in other Galliformes. Eur J Wildl Res 2011;57:983-989.
- Wang YH, Chang H, Xu W, Chang GB. Genetic analysis of microsatellite DNA markers in domestic quail and wild Japanese quail populations. Vcta. Veterinariaet. Zootechnica. Sinica 2004;4:367-371.
- Wu SJ.Evalution of Genetic diversity among four feathers eggs quail populations by using microsatellites.luoyang in China: Master degree thesis of Henan University of Science and Technology,2010.
- Yue GH,Hong LG.Quail yellow feather sexing establish self supporting system - find quail yellow feather recessive sex-linked genes and identification. Beijing: China Agricultural Science and Technology Press, 1994.
Publication Dates
-
Publication in this collection
Jul-Sep 2016
History
-
Received
June 2015 -
Accepted
Feb 2016





 Note: M: markers,1:AD,2, 5:CE, 3, 4: BB, 6, 7, 8, 9, 10: BD
Note: M: markers,1:AD,2, 5:CE, 3, 4: BB, 6, 7, 8, 9, 10: BD