ABSTRACT
The carbohydrate response element-binding protein (ChREBP) is an important nuclear factor that regulates glycolysis and de novo lipogenesis. However, the role of ChREBP in fatty liver development in geese remains unclear. In order to understand the function of ChREBP in lipid metabolism of geese, we first cloned the complete cDNA of the ChREBP of the Sichuan White goose (Anser cygnoides) using RT-PCR, 5’ RACE and 3’ RACE, and analyzed goose ChREBP expression in nine different tissues using real-time PCR technology. The results showed that the goose ChREBP CDS consists of 945bp nucleotides that encode 314 amino acids, and the sequence has high similarities with the swan goose (Anser cygnoides domesticus) and duck (Anas platyrhynchos) sequences, both at the nucleotide and amino acid levels. The predicted ChREBP protein had a molecular mass of 35.64 kDa with pI value of 5.36. The phylogenetic analysis indicated its evolutionary relationships with corresponding orthologous sequences in swan geese and ducks. The qPCR assays revealed that ChREBP is highly expressed in liver in the Sichuan White goose. Together, these results indicate that goose ChREBP may play an important role in the development of hepatic steatosis.
Keywords:
ChREBP; Expression profile; Molecular cloning; RACE; Sichuan White Goose
INTRODUCTION
The transcription factor carbohydrate-responsive element binding protein (ChREBP, also known as MondoB or MLXIPL) is a basic helix-loop-helix/leucine zipper (bHLH/LZ) transcription factor that is predominately expressed in the liver, kidney, and adipose tissue of mammals (Yamashita et al., 2001Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proceedings of the National Academy of Sciences USA 2001;98:9116-9121.; Towle, 2005Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends in Endocrinology and Metabolism 2005;16:489-494.; Filhoulaud et al., 2013Filhoulaud G, Guilmeau S, Dentin R, Girard J, Postic C. Novel insights into ChREBP regulation and function. Trends in Endocrinology & Metabolism 2013;24(5): 257-268.). Its molecular weight is 96 kDa, and it consists of 864 amino acids (Filhouland et al., 2013). This protein was found to bind the carbohydrate response element (ChoRE) and to be involved in the development of metabolic syndromes and glucose-inducible expression of several genes, including fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC), stearoyl-CoA desaturase-1 (SCD1), thioredoxin-interacting protein (TXNIP), and fibroblast growth factor 21 (FGF21) (Katsurada et al., 1990Katsurada A, Iritani N, Fukuda H, Matsumura Y, Nishimoto N, Noguchi T, et al. Effects of nutrients and hormones on transcriptional and post-transcriptional regulation of acetyl-CoA carboxylase in rat liver. European Journal of Biochemistry 1990;190(2):435-41.; Lizuka et al., 2008; Cha-Molstad et al., 2009Cha-Molstad H, Saxena G, Chen J, Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4acetylation in pancreatic beta cells. Journal of Biology Chemistry 2009;284(25): 16898-16905.; Pang et al., 2009Pang ST, Hsieh WC, Chuang CK, Chao CH, Weng WH, Juang HH. Thioredoxin-interacting protein: an oxidative stress-related gene is upregulated by glucose in human prostate carcinoma cell. Journal of Molecular Endocrinology 2009;42(3):205-214.; Jeong et al., 2011Jeong YS, Sim D, Lee YS, Kim HJ, Han JY, Im SS, et al. Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. Plos One 2011;6(7):e22544.; Zhang et al., 2015Zhang GH, Lu JX, Chen Y, Guo PH, Qiao ZL, Feng RF, et al. ChREBP and LXRa mediate synergistically lipogenesis induced by glucose in porcine adipocytes. Gene 2015;565:30-38.). Several lines of evidence suggest that ChREBP interacts with Max-like protein X (Mix) to form a heterodimer, and this ChREBP/Mix heterodimer binds to ChoRE for the activation of the ChoRE-containing promoters in response to high glucose (Stoeckman et al., 2004Stoeckman AK, Ma L, Towle HC. Mix is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. Journal of Biological Chemistry 2004;279:15662-15669.; Ma et al., 2005; Uyeda et al., 2006Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metabolism 2006;4(2):107-110.).
In addition, a recent study reported that ChREBP contains several phosphorylation sites for cAMP-dependent protein kinase (PKA) and AMP-activated protein kinase (AMPK), and it is likely to be regulated by these kinases (Kawaguchi et al., 2001Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphrylation/dephosphorlation of the carbohydrate response element binding protein. Proceedings of the National Academy of Sciences USA 2001;98:13710-13715.). In addition, the phosphorylation of amino acid residues, including Ser 196, Ser 626, Thr666 and Ser568, determines the cellular localization and DNA binding ability of ChREBP (Kawaguchi et al., 2001, 2002). On the contrary, high glucose levels in liver prompt the activation of protein phosphatase 2A (PP2A) through formation of the intermediate of the pentose phosphate shunt, xylulose-5-phosphate (X5P), which dephosphorylates ChREBP, rendering it active (Dentin et al., 2004Dentin R, Pegorier JP, Benhamed F, Foufelle F, Ferre P, Fauveau V, et al. Hepatic blucokinase is iequired for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. Journal of Biological Chemistry 2004;279:20314-20326.). In addition to this “classical” model of ChREBP regulation, a new isoform of ChREBP, named ChREBP-b, plays an important role in the regulation of fatty acid synthesis in adipose tissues and of insulin sensitivity (Herman et al., 2012Herman MA, Peroni OD, Villoria J, Schön MR, Abumrad NA, Blüher MA. Novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 2012;484(7394):333-338.). Recent studies have shown that ChREBP activity can also be regulated by other post-translational modifications, including acetylation and O-linked GlcNAcylation (Sakiyama et al., 2010Sakiyama H, Fujiwara N, Noguchi T, Eguchi H, Yoshihara D, Uyeda K, et al. The role of O-linked GlcNac modification on the glucose response of ChREBP. Biochemical and Biophysical Research Communications 2010;402(4):784-789.; Guinez et al., 2011Guinez C, Filhoulaud G, Rayah-Benhamed F, Marmier S, Dubuquoy C, Dentin R, et al. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 2011;60:1399-1413.). In summary, the tissue distribution, regulation, and DNA-binding properties of ChREBP/MIx suggest that the functions of ChREBP are more complicated than expected.
However, to our knowledge, almost all of the studies on ChREBP have focused on its functions on the lipogenesis of humans, mice, and pigs. By contrast, there are no studies on the role of ChREBP in hepatic steatosis of geese. Therefore, in current study, the full-length cDNA of Sichuan White goose (Anser cygnoides) ChREBP was obtained by rapid amplification of cDNA ends (RACE), and its expression profile in different tissues was determined using real-time PCR. This fundamental work may provide insight into the role of ChREBP in hepatic steatosis, and its theoretical basis could be applied in future research studies.
MATERIALS AND METHODS
Experimental animals and tissue collection
The Sichuan White goose (Anser cygnoides) is a native breed of the Sichuan Province, China. The birds were obtained from the Experimental Farm for Waterfowl Breeding of Sichuan Agricultural University. Five male Sichuan White geese (Anser cygnoides), with 30±3 days of age and of 400±50 g body weight, were randomly selected and sacrificed. For cloning and expression analysis, various tissue samples, including breast muscle, thigh muscle, brain, testis, heart, small intestine, liver, kidney, abdominal fat and sebum cutaneum were collected immediately after sacrifice, and stored in liquid nitrogen at -80ºC for later analyses.
All experimental procedures followed the policies and guidelines of the Sichuan Agricultural University Animal Care and Use Committee (Permit No: 2014-18).
Total RNA extraction and reverse transcription
Total RNA was isolated from the collected tissues using Trizol Reagent (Invitrogen, USA), according to the manufacture’s protocol. The extracted RNA was dissolved in 20-30µL of RNA-free water, and stored at -80°C until thawed for reverse transcription. The integrity of the RNA was evaluated by electrophoresis on 1.0% agarose gel, and RNA concentration and purity were detected using a spectrophotometer at a 260/280 nm absorbance ratio (NanoVue PlusTM, Thermo Scientific, USA). Reverse transcription of total RNA was carried out using the PrimeScript TMRT reagent kit (TaKaRa, Dalian, China), according to the manufacturer’s instructions. The reaction was performed in a total volume 10 µL containing 2 µL of 5×PrimeScriptTM buffer, 0.5 µL of PrimeScriptTM RT Enzyme Mix I, 0.5 µL of Oligo dT Primer, 0.5 µL of random hexamers, 5.5 µL of RNase-free water, and 1 µg of total RNA. The reaction conditions for cDNA synthesis were 37°C for 15 min, followed by 85°C for 5 s, and storage at 4°C. The cDNA was directly used in quantitative real-time PCR.
Cloning and sequencing of the full-length cDNA of Sichuan White goose (Anser cygnoides) ChREBP gene
According to the suppression subtractive hybridization (SSH) results described in our previous report (Pan et al., 2011Pan ZX, Han CC, Wang JW, Li L, Tang H, Lv J, et al. Cloning and expression of stearoyl-CoA desaturase 1 (SCD-1) in the liver of the Sichuan white goose and landes goose responding to overfeeding. Molecular Biololgy Reports 2011;38:3417-3425.), we found that an EST of 481 bp (GW342987), which was homologous to the ChREBP of other species, according to BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/). Therefore, this EST sequence was selected for further cloning of the full-length cDNA of Sichuan White goose chREBP gene. According to the EST sequence of ChREBP, two specific primers (ChREBPGSP1 and ChREBPGSP2) were designed to carry out 5’ and 3’-RACE. ChREBPGSP1 was used to amplify the 5’ end, and chREBPGSP2 was used to amplify the 3’ end. The univeral primer (UPM) used for 5’ and 3’-RACE was the mixture of the Long primer with the Short Primer (from SMARTTM RACE cDNA amplification kit, Clontech, USA). The above primers are listed in Table 1. The amplification of the 5’ and 3’ termini of the ChREBP from Sichuan White goose liver by 5’-RACE and 3’-RACE was performed using the SMART TM RACE cDNA amplification kit (Clontech, USA), according to the manufacturer’s instructions. Touchdown PCR was used for RACE amplification as follows: 4 min at 94°C; five cycles of 35 s at 94°C, 3 min at 72°C; five cycles of 35 s at 94°C, 3 min at 72°C; 25 cycles of 35 s at 94°C, 30 s at 68°C, and 3 min at 72°C, followed by an elongation step of 10 min at 72°C. The PCR products of 5’ and 3’ RACE were separated by electrophoresis on 3% agarose gel and purified using the E.Z.N.A.® Gel Extraction Kit (Omega Bio-Tek, Noraville, GA, USA). The purified fragments were cloned into the pMD-18T vector (Takara, Dalian, Liaoning, China), and five to ten clones were selected for sequencing.
Bioinformatic analysis of ChREBP
The obtained nucleotide sequences were analyzed at NCBI (http://blast.ncbi.nlm.nih.gov/) and were compared against the sequence database using the BLAST server (http://www.ncbi.nlm.nih.gov/blast). The Prot Param tool (http://www.expasy.org/tools/protparam.html) was used to analyze the fundamental characteristics of the predicted proteins, including isoelectric point (pI) and molecular weight (Mw). The conserved domain of the protein was analyzed using the CD-search tool on NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). The secondary structure was predicted by the NPS@ service (http://npsa-pbil.ibcp.fr/). The amino acid sequence was analyzed for transmembrane structure via TMHMM server (http://www.cbs.dtu.dk/services/TMHMM) and signal protein prediction via SignaIP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/). NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc/) and NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) were used to predict potential O- and N-glycosylation sites, respectively. Protein phosphorylation sites were predicted using NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/). Multiple alignments of ChREBP amino acid sequences of the Sichuan white goose with other species were performed using the DNAMAN 6.0, and the phylogenetic tree was constructed using Molecular Evolutionary Genetic Analysis (MEGA) software version 5.2 (http://www. megasoftware.net/).
Analysis of the expression of the ChREBP gene
For the gene expression analysis of ChREBP in different tissues, we performed quantitative real-time PCR for ChREBP by using the iCycler IQ5 Multicolor Real-time PCR detection system (Bio-Rad, USA) with the SYBR PrimerScrip RT-PCR kit (Takara, Dalian, China). The primers of ChREBP and two housekeeping internal control genes (18s and b-actin) were shown in Table 1. Real-time PCR reactions were carried out in a volume of 25 µL, which contained 2.0 µL of cDNA, 0.5 µL of each primers, 12.5 µL of SYBR Premix EX TaqTM and 9.5 µL of water. The procedure included one cycle of 95°C for 10 s, followed by 40 cycles of 95°C for 10 s and 60°C for 20 s. A 45-cycle melting curve was performed, starting at a temperature of 55°C and increasing by 0.5°C every 10s to determine primer specificity. Each sample was repeated in triplicate. The relative gene expression levels were normalized to 18s, b-actin and further expressed as fold change relative to the expression level in control using 2-△△CT method in the gene expression assay.
Statistical analysis
Variations were analyzed via one-way analysis of variance using SAS 6.12 software, a multiple comparison test was performed using Duncan’s method, and p<0.05 was considered statistically significant. The means ± SEM results were plotted to the figures using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA).
RESULTS
Cloning and analysis of full-length ChREBP from Sichuan White goose (Anser cygnoides)
Using degenerate primers and RACE strategy, we cloned ChREBP from the liver tissue of the Sichuan White goose. All obtained fragments were assembled using the BioEdit software to yield a full-length goose ChREBP cDNA with 1262 bp. Sequence analysis using the ExPASy translate toll showed that the sequence contained a 125-bp 5’ untranslated region (5’ UTR), a 192-bp 3’ untranslated region (3’UTR) with a 22 bp poly (A) tail, and an open reading frame (ORF) of 945-bp encoding 314 amino acids with a predicted molecular weight (Mw) of 35.64 kDa, and a isoelectric point (Ip) of 5.36 (Fig.1).
Full-length of Sichuan White goose (Anser cygnoides) ChREBP cDNA sequence and the deduced amino acid sequence. The stop codon is shown as “*”. Numbers on the left refer to the nucleotide position.
Characterization of Sichuan White goose (Anser cygnoides) ChREBP
The analysis using NetPhos 2.0 predicted a total of 17 phosphorylation sites, which consisted of 14 Ser phosphorylation sites (Ser23, 27, 29, 52, 62, 64, 82, 104, 153, 196, 197, 203, 204 and 292), 2 Thr phosphorylation sites (Thr148 and 215), and 1 Tyr phosphorylation site (Tyr275) of goose ChREBP. On the other hand, the Sichuan White goose ChREBP was predicted to have 1 O-glycosylation site (Thr33), but no N-glycosylation sites. The NCBI conserved domain tool shows that the Sichuan White goose ChREBP does not contain a conservative field. The secondary structure prediction of Sichuan White goose ChREBP protein was deduced by NPS@ service based on the HNNC, MLRC and PHD methods. The 2D structure of Sichuan White goose ChREBP protein presented average alpha helices and abundant random coil, and less extended strands. Beta turn and other rare secondary structures were absent (Table 2). No signal peptide and transmembrane domain were identified.
Multiple sequence alignment and phylogenetic relationship
The nucleotide sequence and the deduced amino acid sequences of the Sichuan White goose ChREBP gene were aligned with those of other species using the DNAMAN listed in Table 3. The amino acid alignment of these species shown that there are high similarities in ChREBP gene between goose and other birds, therefore, ChREBP may be conserved protein in birds (Figure 2).
DNAMAN program aligment of the deduced amino acid sequences of the Sichuan White goose (Anser cygnoides) with that of other animal species.
The conserved domains are shown in dark blue.
In order to evaluate the molecular evolutionary relationship of ChREBP in birds and mammals, a phylogenetic tree was constructed using 17 ChREBP protein sequences from various animals by the neighbor-joining method. As shown in Fig. 3, the ChREBP genes of these species were divided into two groups representing mammals and birds, with Sichuan White goose ChREBP belonging to the bird cluster, thus closely related to Anser cygnoides domesticus, Anas platyrhynchos and Gallus gallus.
Phylogenetic tree illustrating the genetic relationships of the ChREBP of Sichuan White goose (Anser cygnoides) with those of other animals. Numbers on each branch indicate the percentage of times a node was supported in 1000 bootstrap pseudo replications by neighbor joining. Sequence name and accession number in the figure are shown as followed: Anser cygnoides domesticus (XP_013044706.1), Anas platyrhynchos (XP_012951552.1), Gallus gallus (NP_001104311.1), Charadrius vociferus (XP_009887195.1), Manacus vitellinus (XP_017931999.1), Haliaeetus leucocephalus (XP_010584275.1), Calidris pugnax (XP_014815208.1), Columba livia (XP_013224200.1), Egretta garzetta (XP_009643245.1), Parus major (XP_015502361.1), Equus przewalskii (XP_008536526.1), Monodelphis domestica (XP_016285149.1), Chelonia mydas (XP_007069093.1), Rattus norvegicus (NP_598236.1), Mus musculus (NP_067430.2), and Homo sapiens (NP_116572.1). The scale bar indicates the branch length.
Expression pattern of ChREBP
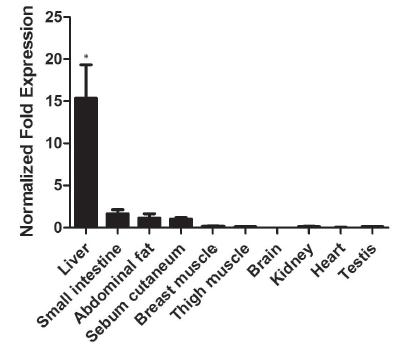
For the purpose of clarifying the expression of the ChREBP gene in each tissue, we used 18s and b-actin as the internal reference and brain as the control. The ChREBP gene was found to be expressed in all tissues, as shown in Figure 4. However, its expression level varied significantly. Based on the fold changes relative to the brain, the Sichuan White goose ChREBP mRNA was over 15-fold more abundant in the liver, whereas its expression was low in the abdominal fat, sebum cutaneum, and small intestine, and almost undetectable in the brain, testis, kidney, heart, and thigh and breast muscle tissues. Therefore, the influence of ChREBP probably depends on the function of the different tissues.
Relative mRNA expression of the ChREBP gene in different tissues, including liver, small intestine, abdominal fat, sebum cutaneum, breast muscle, thigh muscle, heart, kidney, brain and testis. Data are presented as mean ± SEM (n=3). Error bars show the SEM of triplicate. “*” indicates significant difference (p<0.05).
DISCUSSION
ChREBP is an important glucose-responsive transcription factor, and affects various biological processes, such as glucolipotoxicity to apoptosis (Reich et al., 2012Reich E. Thioredoxin-interacting protein mediates ER stress-induced beta cell death through initiation of the inflammasome. Cell Metabolism 2012;16:265-273.; Filhoulaud et al., 2013Filhoulaud G, Guilmeau S, Dentin R, Girard J, Postic C. Novel insights into ChREBP regulation and function. Trends in Endocrinology & Metabolism 2013;24(5): 257-268.), and proliferation of specific cell types (Metukuri et al., 2012Metukuri M. ChREBP mediateds glucose-stimulated pancreatic b-cell proliferation. Diabetes 2012;61:2004-2015.; Soggia et al., 2012Soggia A, Flosseau K, Ravassard P, Szinnai G, Scharfmann R, Guillemain G. Activation of the transcription factor ChREBP by glucose leasds to increased pancreatic b-cell differentiation in rats. Diabetologia 2012;55:2713-2722.). To date, ChREBP has been identified and characterized in humans (Hurtado del Pozo et al., 2011Hurtado del Pozo C, Vesperinas-García G, Rubio MÁ, Corripio-sánchez R, Torres-García AJ, Obergon MJ, et al. ChREBP expression in the liver, adipose tissue and differentiated preadipocytes in human obesity. Biochimica Biophysica Acta 2011;1811(12):1194-1200.; Stamatikos et al., 2015Stamatikos AD, da Silva RP, Lewis JT, Donglas DN, Kneteman NM, Jacobs RL, et al. Tissue specific effects of dietary carbohydrates and obesity on ChREBPa and ChREBPb expression. Lipids 2015;2:1-10.) and mice (Dentin et al., 2006Dentin R, Benhamed F, Hainault I, Fauveau V, Foufelle F, Dyck JR. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes 2006;55(8): 2159-2170.). However, no previous studies have been reported describing the structure of ChREBP gene in non-mammalian vertebrates, including birds.
In this study, we cloned ChREBP cDNA from the liver tissue of Sichuan White geese. The total Sichuan White goose ChREBP CDS consists of 945 nucleotides that encode 314 amino acids. These results are different from those of a previous study showing that ChREBP is a large protein containing 864 amino acids in humans (Filhoulaud et al., 2013Filhoulaud G, Guilmeau S, Dentin R, Girard J, Postic C. Novel insights into ChREBP regulation and function. Trends in Endocrinology & Metabolism 2013;24(5): 257-268.). The deduced amino acid sequence of Sichuan White goose ChREBP was highly similar to those of other species, particularly of swan geese and ducks (more than 94%). The results of ChREBP phylogenetic relationship also identified the Sichuan White goose ChREBP in nearly all bird species examined, especially in domestic geese (Anser cygnoidest domesticus), mallards (Anas platyrhynchos) and domestic chickens (Gallus gallus). This result was consistent with their zoological classification and indicated that this protein was highly evolutionarily conserved. As in mammalian vertebrates, the ChREBP protein identified contains several key domains, including a nuclear localization signal (NLS), polyproline domains, a bHLH/LZ domain, and a leucine zipper-like (Zip-like) domain (Filhoulaud et al., 2013; Schmidt, et al., 2016Schmidt SF, Maden JG, Frafjord KØ, Poulsen LI, Salö S, Boergesen M, et al. Integrative genomics outlines a biphasic glucose response and a ChREBP-RORg axis regulating proliferation in b cells. Cell Reports 2016;16:2359-2372.); however, no conservative domain was detected in the Sichuan White goose. The different results might be due to differences among species. Moreover, other structural features, such as Ser 196 at one of the phosphorylation sites of goose ChREBP is consistent with the ChREBP gene in other species, which regulates nuclear localization (Tsatsos et al., 2008Tsatsos N. Identification and function of phosphorylation in the glucose-regulated transcription factor ChREBP. Biochemical Journal 2008;411:261-270.; Krivoruchko et al., 2014Krivoruchko A, Storey KB. Activation of the carbohydrate response element binding protein (ChREBP) in response to anoxia in the turtle Trachemys scripta elegans. Biochimica et Biophysica Acta 2014;1840:3000-3005.).
In mammals, ChREBP are expressed in all types of tissues, with high expression level in the liver, kidney, and adipose tissue (Towle, 2005Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends in Endocrinology and Metabolism 2005;16:489-494.). In mice, the ChREBP mRNA is first detected in the liver at embryonic stage E12.5 (Cairo et al., 2001Cairo S, Merla G, Urbinati F, Ballabio A, Reymond A. WBSCR 14, a gene mapping to the Williams-Beuren syndrome deleted region, is a new member of the Mix transcription factor network. Human Molecular Genetics 2001;10:617-627.), and in adulta, ChREBP is primarily expressed in the liver and the adipose tissue, with low expression levels in the intestine, kidney, and muscle (Iizuka et al., 2004Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proceedings of the National Academy of Sciences USA 2004;101:7281-7286.; Yamashita et al., 2001Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proceedings of the National Academy of Sciences USA 2001;98:9116-9121.). In the current study, to better understand the expression pattern of the Sichuan White goose ChREBP gene, the its mRNA level was analyzed in nine different tissues. The results showed that the ChREBP gene was mainly expressed in the liver and the abdominal fat, which is consistent with previously reported data for the differential tissue expression of ChREBP in mammals (Towle, 2005), suggesting that ChREBP has a important role in the regulation of glycolytic reaction and lipogenesis. However, the detailed mechanisms by which ChREBP function in these specific tissues remain to be elucidated.
In summary, a cDNA clone of ChREBP was the first to be cloned and characterized in the Sichuan White goose. Sequence analysis showed that Sichuan White goose (Anser cygnoides) ChREBP has high similarities with those of the swan goose (Anser cygnoides domesticus) and the mallard (Anas platyrhynchos) sequences at both the nucleotide and amino acid levels. Real-time PCR assay revealed that, although ChREBP is widely expressed in nearly all Sichuan White goose tissues examined, its highest expression level was detected in the liver. Taken together, our data suggests that ChREBP may play vital role in hepatic steatosis; however, a larger scale association study involving the molecular mechanism of ChREBP in the regulation of fatty liver development is required to support and substantiate this study.
ACKNOWLEDGMENTS
This work was financially supported by the China Agricultural Research System (CARS-42-4), Key Technology Support Program of Sichuan Province (2016NYZ0027) and A Project Supported by Scientific Reserch Fund of Sichuan Provincial Education Department (15ZA0025).
REFERENCES
- Cairo S, Merla G, Urbinati F, Ballabio A, Reymond A. WBSCR 14, a gene mapping to the Williams-Beuren syndrome deleted region, is a new member of the Mix transcription factor network. Human Molecular Genetics 2001;10:617-627.
- Cha-Molstad H, Saxena G, Chen J, Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4acetylation in pancreatic beta cells. Journal of Biology Chemistry 2009;284(25): 16898-16905.
- Dentin R, Pegorier JP, Benhamed F, Foufelle F, Ferre P, Fauveau V, et al. Hepatic blucokinase is iequired for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. Journal of Biological Chemistry 2004;279:20314-20326.
- Dentin R, Benhamed F, Hainault I, Fauveau V, Foufelle F, Dyck JR. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes 2006;55(8): 2159-2170.
- Filhoulaud G, Guilmeau S, Dentin R, Girard J, Postic C. Novel insights into ChREBP regulation and function. Trends in Endocrinology & Metabolism 2013;24(5): 257-268.
- Guinez C, Filhoulaud G, Rayah-Benhamed F, Marmier S, Dubuquoy C, Dentin R, et al. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 2011;60:1399-1413.
- Herman MA, Peroni OD, Villoria J, Schön MR, Abumrad NA, Blüher MA. Novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 2012;484(7394):333-338.
- Hurtado del Pozo C, Vesperinas-García G, Rubio MÁ, Corripio-sánchez R, Torres-García AJ, Obergon MJ, et al. ChREBP expression in the liver, adipose tissue and differentiated preadipocytes in human obesity. Biochimica Biophysica Acta 2011;1811(12):1194-1200.
- Iizuka K, Horikawa Y. ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocrine Journal 2008;55:617-624.
- Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proceedings of the National Academy of Sciences USA 2004;101:7281-7286.
- Jeong YS, Sim D, Lee YS, Kim HJ, Han JY, Im SS, et al. Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. Plos One 2011;6(7):e22544.
- Katsurada A, Iritani N, Fukuda H, Matsumura Y, Nishimoto N, Noguchi T, et al. Effects of nutrients and hormones on transcriptional and post-transcriptional regulation of acetyl-CoA carboxylase in rat liver. European Journal of Biochemistry 1990;190(2):435-41.
- Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphrylation/dephosphorlation of the carbohydrate response element binding protein. Proceedings of the National Academy of Sciences USA 2001;98:13710-13715.
- Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid "sparing" effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. Journal of Biological Chemistry 2002;277(6):3829-3835.
- Krivoruchko A, Storey KB. Activation of the carbohydrate response element binding protein (ChREBP) in response to anoxia in the turtle Trachemys scripta elegans. Biochimica et Biophysica Acta 2014;1840:3000-3005.
- Ma L, Tsatsos NG, Towle HC. Direct role of ChREBP Mix in regualting hepatic glucose-responsive genes. Journal of Biological Chemistry 2005;280:12019-12027.
- Metukuri M. ChREBP mediateds glucose-stimulated pancreatic b-cell proliferation. Diabetes 2012;61:2004-2015.
- Pan ZX, Han CC, Wang JW, Li L, Tang H, Lv J, et al. Cloning and expression of stearoyl-CoA desaturase 1 (SCD-1) in the liver of the Sichuan white goose and landes goose responding to overfeeding. Molecular Biololgy Reports 2011;38:3417-3425.
- Pang ST, Hsieh WC, Chuang CK, Chao CH, Weng WH, Juang HH. Thioredoxin-interacting protein: an oxidative stress-related gene is upregulated by glucose in human prostate carcinoma cell. Journal of Molecular Endocrinology 2009;42(3):205-214.
- Reich E. Thioredoxin-interacting protein mediates ER stress-induced beta cell death through initiation of the inflammasome. Cell Metabolism 2012;16:265-273.
- Sakiyama H, Fujiwara N, Noguchi T, Eguchi H, Yoshihara D, Uyeda K, et al. The role of O-linked GlcNac modification on the glucose response of ChREBP. Biochemical and Biophysical Research Communications 2010;402(4):784-789.
- Schmidt SF, Maden JG, Frafjord KØ, Poulsen LI, Salö S, Boergesen M, et al. Integrative genomics outlines a biphasic glucose response and a ChREBP-RORg axis regulating proliferation in b cells. Cell Reports 2016;16:2359-2372.
- Stamatikos AD, da Silva RP, Lewis JT, Donglas DN, Kneteman NM, Jacobs RL, et al. Tissue specific effects of dietary carbohydrates and obesity on ChREBPa and ChREBPb expression. Lipids 2015;2:1-10.
- Stoeckman AK, Ma L, Towle HC. Mix is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. Journal of Biological Chemistry 2004;279:15662-15669.
- Soggia A, Flosseau K, Ravassard P, Szinnai G, Scharfmann R, Guillemain G. Activation of the transcription factor ChREBP by glucose leasds to increased pancreatic b-cell differentiation in rats. Diabetologia 2012;55:2713-2722.
- Tsatsos N. Identification and function of phosphorylation in the glucose-regulated transcription factor ChREBP. Biochemical Journal 2008;411:261-270.
- Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends in Endocrinology and Metabolism 2005;16:489-494.
- Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metabolism 2006;4(2):107-110.
- Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proceedings of the National Academy of Sciences USA 2001;98:9116-9121.
- Zhang GH, Lu JX, Chen Y, Guo PH, Qiao ZL, Feng RF, et al. ChREBP and LXRa mediate synergistically lipogenesis induced by glucose in porcine adipocytes. Gene 2015;565:30-38.
Publication Dates
-
Publication in this collection
Oct-Dec 2017
History
-
Received
30 May 2017 -
Accepted
03 July 2017



 The sequences compared are from Anser cygnoides domesticus (XP_013044706.1), Anas platyrhynchos (XP_012951552.1), Gallus gallus (NP_001104311.1), Charadrius vociferus (XP_009887195.1), Manacus vitellinus (XP_017931999.1), Haliaeetus leucocephalus (XP_010584275.1), Calidris pugnax (XP_014815208.1), Columba livia (XP_013224200.1), Egretta garzetta (XP_009643245.1), Parus major (XP_015502361.1), Equus przewalskii (XP_008536526.1), Monodelphis domestica (XP_016285149.1), Chelonia mydas (XP_007069093.1), Rattus norvegicus (NP_598236.1), Mus musculus (NP_067430.2), and Homo sapiens (NP_116572.1).
The sequences compared are from Anser cygnoides domesticus (XP_013044706.1), Anas platyrhynchos (XP_012951552.1), Gallus gallus (NP_001104311.1), Charadrius vociferus (XP_009887195.1), Manacus vitellinus (XP_017931999.1), Haliaeetus leucocephalus (XP_010584275.1), Calidris pugnax (XP_014815208.1), Columba livia (XP_013224200.1), Egretta garzetta (XP_009643245.1), Parus major (XP_015502361.1), Equus przewalskii (XP_008536526.1), Monodelphis domestica (XP_016285149.1), Chelonia mydas (XP_007069093.1), Rattus norvegicus (NP_598236.1), Mus musculus (NP_067430.2), and Homo sapiens (NP_116572.1).