Abstract
BACKGROUND: The Kell blood group system expresses high and low frequency antigens with the most important in relation to transfusion including the antithetic KEL1 and KEL2; KEL3 and KEL4; KEL6 and KEL7 antigens. Kell is a clinically relevant system, as it is highly immunogenic and anti-KEL antibodies are associated with hemolytic transfusion reactions and hemolytic disease of the fetus and newborn. Although required in some situations, Kell antigen phenotyping is restricted due to technical limitations. In these cases, molecular approaches maybe a solution. This study proposes three polymerase chain reaction genotyping protocols to analyze the single nucleotide polymorphisms responsible for six Kell antithetic antigens expressed in a Brazilian population. METHODS: DNA was extracted from 800 blood donor samples and three polymerase chain reaction-restriction fragment length polymorphism protocols were used to genotype the KEL*1/KEL*2, KEL*3/KEL*4 and KEL*6/KEL*7 alleles. KEL*3/KEL*4 and KEL*6/KEL*7 genotyping was standardized using the NlaIII and MnlI restriction enzymes and validated using sequencing. KEL*1/KEL*2 genotyping was performed using a previously reported assay. RESULTS: KEL genotyping was successfully implemented in the service; the following distribution of KEL alleles was obtained for a population from southeastern Brazil: KEL*1 (2.2%), KEL*2 (97.8%), KEL*3 (0.69%), KEL*4 (99.31%), KEL*6 (2.69%) and KEL*7 (97.31%). Additionally, two individuals with rare genotypes, KEL*1/KEL*1 and KEL*3/KEL*3, were identified. CONCLUSION: KEL allele genotyping using these methods proved to be reliable and applicable to predict Kell antigen expressions in a Brazilian cohort. This easy and efficient strategy can be employed to provide safer transfusions and to help in rare donor screening.
Kell blood-group system; Molecular biology; Gene frequency; Erythrocytes; Polymerase chain reaction
ORIGINAL ARTICLE
An easy and efficient strategy for KEL genotyping in a multiethnic population
Carine Prisco ArnoniI; Janaína Guinhem MunizI; Tatiane Aparecida de PaulaI; Rosangela Duarte de Medeiros PersonI; Diana GazitoI; Wilson Baleotti JrII; José Augusto BarretoI; Lilian CastilhoIII; Flavia Roche Moreira LatiniI
IAssociação Beneficente de Coleta de Sangue - COLSAN, São Paulo, SP, Brazil
IIFaculdade de Medicina de Marilia - FAMEMA, Marília, SP, Brazil
IIIUniversidade Estadual de Campinas - UNICAMP, Campinas, SP, Brazil
Corresponding author Corresponding author: Carine Prisco Arnoni Associação Beneficente de Coleta de Sangue - COLSAN Av. Jandira, 1260 Indianópolis 04080-006 São Paulo, SP, Brazil carine.arnoni@colsan.org.br
ABSTRACT
BACKGROUND: The Kell blood group system expresses high and low frequency antigens with the most important in relation to transfusion including the antithetic KEL1 and KEL2; KEL3 and KEL4; KEL6 and KEL7 antigens. Kell is a clinically relevant system, as it is highly immunogenic and anti-KEL antibodies are associated with hemolytic transfusion reactions and hemolytic disease of the fetus and newborn. Although required in some situations, Kell antigen phenotyping is restricted due to technical limitations. In these cases, molecular approaches maybe a solution. This study proposes three polymerase chain reaction genotyping protocols to analyze the single nucleotide polymorphisms responsible for six Kell antithetic antigens expressed in a Brazilian population.
METHODS: DNA was extracted from 800 blood donor samples and three polymerase chain reaction-restriction fragment length polymorphism protocols were used to genotype the KEL*1/KEL*2, KEL*3/KEL*4 and KEL*6/KEL*7 alleles. KEL*3/KEL*4 and KEL*6/KEL*7 genotyping was standardized using the NlaIII and MnlI restriction enzymes and validated using sequencing. KEL*1/KEL*2 genotyping was performed using a previously reported assay.
RESULTS: KEL genotyping was successfully implemented in the service; the following distribution of KEL alleles was obtained for a population from southeastern Brazil: KEL*1 (2.2%), KEL*2 (97.8%), KEL*3 (0.69%), KEL*4 (99.31%), KEL*6 (2.69%) and KEL*7 (97.31%). Additionally, two individuals with rare genotypes, KEL*1/KEL*1 and KEL*3/KEL*3, were identified.
CONCLUSION: KEL allele genotyping using these methods proved to be reliable and applicable to predict Kell antigen expressions in a Brazilian cohort. This easy and efficient strategy can be employed to provide safer transfusions and to help in rare donor screening.
Keywords: Kell blood-group system; Molecular biology; Gene frequency; Erythrocytes; Polymerase chain reaction
Introduction
The Kell blood group system is the third most polymorphic known to date and one of the most clinically relevant in respect to triggering immune reactions. The Kell blood group system comprises at least 36 antigens(1) that are carried on a single red-cell transmembrane protein of 93 kDa(2). Most of the amino acid residues responsible for displaying different Kell antigens are present in the non-conserved globular domain that is away from the plasma membrane. A few of the amino acid substitutions occur in the conserved domain but they are also on the protein surface(3).
Some of Kell antigens are arranged in antithetical set pairs of high and low frequency antigens and others are independently expressed antigens, for which antithetical partners have not been found(4, 5). The antithetical antigens include KEL1 (K, "Kell") and KEL2 (k, "Cellano"); KEL3 (Kpª), KEL4 (Kpb) and KEL21 (Kpc); and KEL6 (Jsª) and KEL7 (Jsb)(5). KEL4, unlike the other high-prevalence antigens, is associated with two antithetical low-prevalence antigens, KEL3 and KEL21(6). Individuals who are negative for a high frequency antigen, including KEL:-2, KEL:-4 or KEL:-7, present rare phenotypes. Weakened Kell antigens may be found in individuals with absent XK protein expressions (McLeod phenotype) or some Gerbich-negative phenotypes(7, 8). Another rare variant phenotype associated with the Kell blood group system is Ko(null), which lacks all Kell antigens and displays enhanced Kx antigens(9).
The antigens are derived from single nucleotide polymorphisms (SNPs) in the KEL gene, which is located at 7q33 and contains 19 exons(10). KEL1 and KEL2 antigens result from a SNP (C578T) in exon 6 that produces a T193M amino acid change. KEL3 and KEL4 antigens result from a point mutation in exon 8 (C841T) that leads to a tryptophan in KEL3 instead of an arginine in KEL4 at amino acid position 281. KEL6 and KEL7 antigens are related to a SNP in exon 17 (T1790C) that encodes a proline in KEL6 or a leucine in KEL7(1).
Antibodies against antigens in the Kell blood group system are usually immunoglobulin G, that can cause severe hemolytic transfusion reactions, as well as hemolytic disease of the fetus and newborn (HDFN). The most important is anti-KEL1, which is a clinically significant antibody. HDFN used to be most commonly associated to Rh alloimmunization, but the use of anti-RhD immunoglobulin as a prophylactic agent has decreased this, and, consequently, HDFN caused by anti-KEL is now more frequent. Anti-KEL1 currently accounts for approximately 10% of the cases of severe anemia in newborns(11). Furthermore, this antibody has already been reported in the induction of myelosuppression, which probably contributes to the anemia(12). Although observed at a much lower frequency, anti-KEL2(13), anti-KEL3(14), anti-KEL4(15), anti-KEL6(16) and anti-KEL7(17) have also been correlated with moderate to severe HDFN.
Antigen frequencies vary in populations from different ethnic backgrounds. Differences in the frequencies of red blood cell (RBC) antigens between European and African descendants have great importance in transfusion medicine, mainly in a multiethnic population. For example, a patient of African origin with a KEL:6, -7 phenotype may be transfused with blood from donors of European origin. As a result, this patient may produce anti-KEL7; when future transfusions are required in these cases, KEL:6, -7 RBCs are necessary(6, 18, 19). Even though the frequency of this phenotype is very low, the identification of KEL6 and KEL7 may be difficult as there is a lack of commercial antibodies and specific and potent antis era are not readily available(20). Besides reagent limitations, phenotyping may also be impaired in other situations, such as when a patient has recently been transfused or has hemolytic anemia or when large-scale typing is required(21).
Considering the importance of Kell antigens in alloimmunization and the limitations of serologic methods, this study reports on the use of a previously reported assay forKEL*1/KEL*2 genotyping(22) and the development of polymerase chain reaction (PCR) restriction fragment length polymorphism (RFLP) techniques to KEL*3/KEL*4 and KEL*6/KEL*7 alleles.
Methods
Blood samples and DNA extraction
Eight hundred blood samples were selected from volunteer blood donors at the Associação Beneficente de Coleta de Sangue (COLSAN). All donors gave their informed consent and 200-µL blood samples were used for DNA extraction with the DNA blood mini kit (QIAamp, Qiagen, Inc., Valencia, CA) following the manufacturer's instructions. DNA concentration was estimated using the NanoDrop 2000 Spectrophotometer (Thermal Cycler, Uniscience Inc., São Paulo, SP, Brazil) and DNA samples were kept at -20ºC for long-term storage.
Polymerase chain reaction primer design and amplification
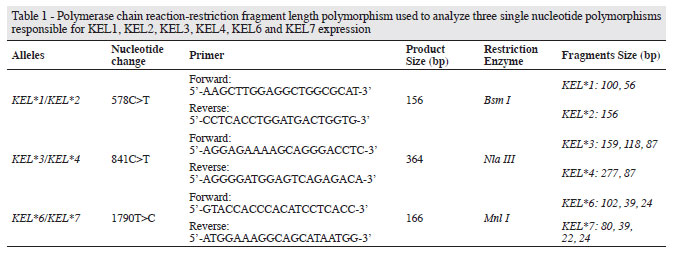
The KEL gene was selected in the ensemble database (http://www.ensembl.org/index.html) and primers were designed using the Primer 3 program (http://frodo.wi.mit.edu/). Hairpin and autodimer formation were evaluated using Autodimerv1removal (http://www.cstl.nist.gov/biotech/strbase/AutoDimerHomepage/AutoDimerProgramHomepage.htm)(23). Alleles, nucleotide changes and primer sequences are described in Table 1.
Polymerase chain reaction (PCR) was performed with 100 ng of DNA, 10 pmol of each primer, 5 nmol of each dNTP, 50 nmol of MgCl2, 1.0 U Taq DNA polymerase and buffer in a final volume of 25 µL. The PCR sequence used in all assays was as follows: 15 min at 95ºC; 35 cycles of 30 s at 94ºC, 30 s at 62ºC and 30 s at 72ºC; followed by 10 min at 72ºC.
Polymerase chain reaction-restriction fragment length polymorphism analysis
After amplification, 5 µL of PCR product was digested for 8 hours at 37ºC with the appropriate restriction enzyme, according to Table 1. Restriction enzyme digestion was performed in a final volume of 10 µL under the conditions recommended by the manufacturer. RFLP bands were analyzed after electrophoresis in 2% agarose gel for KEL*1/KEL*2 and KEL*3/KEL*4 genotyping and in 4% agarose gel for KEL*6/KEL*7 genotyping. Gels were stained with GelRedTM nucleic acid gel stain (Biotium, Inc, Hayward, CA), 10, 000x in water.
Polymerase chain reaction-restriction fragment length polymorphism validation
To check the accuracy of the protocol used, ten DNA samples, previously genotyped by PCR-RFLP, were sequenced. PCR products amplified as described above were purified with 10units of exonuclease I and 1 unit of FastAPTM Thermosensitive Alkaline Phosphatase (Fermentas, Hanover, MD) at 37ºC during45 min followed by heating inactivation of enzymes at 80ºC for15 min. Purified PCR products were submitted to sequencing reaction that consisted of 3 µL of purified PCR product, 2 µL of BigDye Terminator v3.1 (Applied Biosystems, Foster City, CA) and an appropriate buffer. The reaction was submitted to 26 cycles at 96ºC for 10 s, 50ºC for 15 s and 60ºC for 4 min. The product was purified using the BigDye X-terminator kit (Applied Biosystems) and sequencing analysis was performed in a 3500xL Genetic Analyzer (Applied Biosystems). Electropherograms were analyzed using sequencing analysis software (Applied Biosystems).
Results
All three assays were standardized in order for them to be performed under the same cycling conditions to optimize time and equipment. It is important to note that digestion time is a critical point to avoid partial digestion that might lead to an incorrect result. PCR and RFLP products are shown in Figure 1. Genotyping was clearly identified, as the fragments are at a safe distance avoiding misinterpretation. Ten samples with different genotypes were sequenced to evaluate the efficiency of the protocol used. All results were compatible with PCR-RFLP (data not shown).
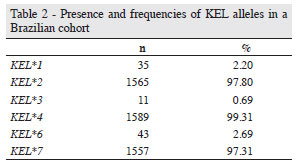
Allele frequencies are shown in Table 2. Of the 800 samples analyzed, the KEL2 allele frequency was 97.8% and one donor with the KEL*1/KEL*1 genotype was identified. A very high frequency of KEL*4 (99.31 %) was observed, however one donor with the rare KEL*3/KEL*3 genotype was also identified. Although the KEL*6 allele was present in a higher frequency of samples thanKEL*3, no donor with the KEL*6/KEL*6 genotype was found.
Discussion
By using PCR-RFLP, a conventional molecular technique, this study combined a previously reported assay to genotype KEL*1/KEL*2(22) with new genotyping assays for the identification of KEL*3/KEL*4 and KEL*6/KEL*7 to successfully analyze 800 DNA samples. Besides reduced costs of PCR-RFLP, these protocols proved to be simple and it seems that any transfusion service, regardless of the size and complexity, could easily implement the technique.
Moreover, these new assays are consistent given that the results from PCR-RFLP analysis were confirmed by sequencing in all the ten subjects used for validating the technique. Through results obtained from the analysis of these three assays, the allele frequencies of the KEL1, KEL2, KEL3, KEL4, KEL5 and KEL6 antigens were characterized in a Brazilian population.
To the best of our knowledge, this is the first report of the frequencies of these alleles in a Brazilian population. The results of this study gave a lower frequency of KEL1 (2.2%) compared to English blood donors (9%). In contrast, the population of the current study presented a frequency of KEL1 higher than in Japanese (0.02%) and Black American populations (1.5%)(18). Although investigated using different strategies, a very low incidence of KEL*3 (0.69%) was observed compared to a previous study that evaluated the frequency of KEL3 (2.28%) in almost 19, 000 Caucasian individuals from Europe and North America using anti-KEL3(19). A higher frequency of KEL*6 (2.69%) was observed in the current study compared to a Caucasian population that was found to be almost 100% negative for KEL6, however it was lower than studies with Afro-American individuals which reported a frequency of 19.5%(6). These results show the ethnic admixture in the Brazilian population, one of the most heterogeneous in the world, a characteristic attributed to the waves of immigration during the colonization process. The frequencies observed in this study reflect the presence of African genetic inheritance, with approximately 3.5 million Africans being introduced to Brazil due to the slave trade after the middle of the 15th century(24). It is essential to consider that the background of the Brazilian population is very distinct depending on the region of the country. The donors in this study are from the southeastern region of Brazil; a region that receives individuals from other regions of the country, especially from the Northeast. Thus, these results may show the mixed allele frequencies of the Brazilian population and not exclusively of this region.
It is also important to emphasize that the assay developed to identify KEL*6/KEL*7 is reliable because the SNP identified as 1790T>C was chosen for genotyping instead of the 2019A>G SNP. High-through put RBC antigen genotyping assays use duplicate probes to detect the same allele with both SNPs, but Renoud et al.(25) demonstrated that probes recognizing KEL at 2019 bp cannot be used to confirm KEL*6/KEL*7 genotyping results. These authors showed that in some Afro-American people the gene encoding the KEL7 antigen is different to the one that was originally described, presenting a cytosine at the 1790 bp position and guanine instead of an adenine in the 2019 position.
Genotyping is an important tool in transfusion medicine and the development and implementation of molecular techniques in blood centers is essential to improve the service and to reduce limitations associated to conventional serology. Kell is an important blood group system and its genotyping allows the prediction of RBC phenotypes when the serology typing is impaired or when it is impossible due to the cost or lack of commercial antis era, as is the case with -KEL3, -KEL4, -KEL5, -KEL6 and -KEL7. Moreover, genotyping allows the identification of rare blood as this study found KEL*3/KEL*3and KEL*1/KEL*1 individuals, which may be useful when blood units are required for alloimmunized patients with anti-KEL1 and -KEL3 antigens.
Conclusion
DNA typing of Kell blood groups by PCR-RFLP using the assays described in this paper can contribute to the management of transfusions in alloimmunized patients by helping to identify antibodies and by allowing the identification of antigen-negative RBC units. This ensures more accurate selection of compatible donor units and is likely to prevent alloimmunization and reduce the incidence of hemolytic reactions.
Submitted: 6/8/2012
Accepted: 11/9/2012
Conflict-of-interest disclosure: The authors declare no competing financial interest
- 1. International Society of Blood Transfusion.[Internet].Amsterdam: ISBT; 2012 [cited 2012 Jun 8]. Available from: http://www.isbtweb.org
- 2. Redman CM, Avellino G, Pfeffer SR, Mukherjee TK, Nichols M, Rubinstein P, et al. Kell blood group antigens are part of a 93, 000-dalton red cell membrane protein. J Biol Chem. 1986;261(20):9521-5.
- 3. Lee S. The value of DNA analysis for antigens of the Kell and Kx blood group systems. Transfusion. 2007;47(1 Suppl):32S-9S.
- 4. Daniels GL, Anstee DJ, Cartron JP, Dahr W, Henry S, Issitt PD, et al. Terminology for red cell surface antigens. Makuhari Report. Vox Sang. 1996;71(4):246-8.
- 5. Daniels GL, Anstee DJ, Cartron JP, Dahr W, Issitt PD, Jørgensen J, et al. Blood group terminology 1995. ISBT Working Party on terminology for red cell surface antigens. Vox Sang. 1995;69(3):265-79.
- 6. Lee S. Molecular basis of Kell blood group phenotypes. Vox Sang. 1997;73(1):1-11. Erratum in: Vox Sang. 1998;74(1):58.
- 7. Daniels G. The molecular genetics of blood group polymorphism. Hum Genet. 2009;126(6):729-42.
- 8. Westhoff CM, Reid ME. Review: the Kell, Duffy, and Kidd blood group systems. Immunohematology. 2004;20(1):37-49.
- 9. Russo D, Redman C, Lee S. Association of XK and Kell blood group proteins. J Biol Chem. 1998;273(22):13950-6.
- 10. Lee S, Zambas E, Green ED, Redman C. Organization of the gene encoding the human Kell blood group protein. Blood. 1995;85(5):1364 70. Erratum in: Blood. 1996;87(11):4922.
- 11. Lee S, Russo D, Redman CM. The Kell blood group system: Kell and XK membrane proteins. Semin Hematol. 2000;37(2):113-21.
- 12. Vaughan JI, Manning M, Warwick RM, Letsky EA, Murray NA, Roberts IA. Inhibition of erythroid progenitor cells by anti-Kell antibodies in fetal alloimmune anemia. N Engl J Med. 1998;338(12):798-803. Comment in: N Engl J Med. 2000;343(1):72; N Engl J Med. 1998;338(12):830-1.
- 13. Duguid JK, Bromilow IM. Haemolytic disease of the newborn due to anti-k. Vox Sang. 1990;58(1):69.
- 14. Smoleniec J, Anderson N, Poole G. Hydrops fetalis caused by a blood group antibody usually undetected in routine screening. Arch Dis Child Fetal Neonatal Ed. 1994c;71(3):F216-7.
- 15. Gorlin JB, Kelly L. Alloimmunisation via previous transfusion places female Kpb-negative recipients at risk for having children with clinically significant hemolytic disease of the newborn. Vox Sang. 1994;66(1):46-8.
- 16. Donovan LM, Tripp KL, Zuckerman JE, Konugres AA. In: Daniels G. Hemolytic disease of the newborn due to anti-Js a. Transfusion. 1973;13(3):153.
- 17. Stanworth S, Fleetwood P, de Silva M. Severe haemolytic disease of the newborn due to anti-Js(b). Vox Sang. 2001;81(2):134-5.
- 18. Daniels G, editor. Kell and Kx blood group systems. Human Blood Groups. 1995, Oxford: Blackwell Science; 1196. p. 385-420.
- 19. Race RR, Sanger R. Blood groups in man. 6th ed. Oxford: Blackwell Scientific, 1975.
- 20. Jungbauer C. Routine use of DNA testing for red cell antigens in blood centres. Transfus Apher Sci. 2011;45(1):61-8.
- 21. Hillyer CD, Shaz BH, Winkler AM, Reid M. Integrating molecular technologies for red blood cell typing and compatibility testing into blood centers and transfusion services. Transfus Med Rev. 2008;22(2):117-32.
- 22. Reid ME, Rios M, Powell VI, Charles-Pierre D, Malavade V. DNA from blood samples can be used to genotype patients who have recently received a transfusion. Transfusion. 2000;40(1):48-53.
- 23. Vallone PM, Butler JM. AutoDimer: a screening tool for primer-dimer and hairpin structures. Biotechniques. 2004;37(2):226-31.
- 24. Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK, Pereira R, Gusmão L, Amorim A, et al. Assessing individual inter ethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31(2):184-90.
- 25. Renoud KJ, Barracchini K, Byrne KM, Adams S, Pickett A, Caruccio L, et al. KEL6 and KEL7 genotyping with sequence-specific primers. Transfusion. 2006;46(9):1510-4.
Corresponding author:
Publication Dates
-
Publication in this collection
05 June 2013 -
Date of issue
2013
History
-
Received
06 Aug 2012 -
Accepted
11 Sept 2012