Abstracts
The analysis of DNA in clinical samples for a secure diagnostic has become indispensable nowadays. Techniques approaching isolation of high molecular weigth DNA of HPV could lead to efficient amplification and early clinical diagnosis of the virus DNA by PCR (polymerase chain reaction). We describe a fast, non-toxical, efficient and cheap method for DNA isolation of human papilloma virus (HPV) from cervical smears using guanidine (DNAzol solution). A 450 bp DNA band correponding to the late region (L1) of the virus genome was detected by PCR, showing that the DNAzol extraction soluction generated a good viral DNA yield. The electrophoretic pattern after digestion with restriction endonucleases (RFLPs/PCR) revealed the predominance of HPV-16 and HPV-33 in the samples from the State of Alagoas, Brazil.
Human papillomavirus; DNA purification; PCR; RFLP
A detecção de DNA em amostras clínicas visando um diagnóstico mais seguro vem se tornando uma prática comum em laboratórios de análise clínica. Metodologias que objetivem o isolamento de DNA de alto peso molecular de HPV podem levar a uma amplificação precisa e diagnose precoce do DNA do vírus por PCR (reação de polimerase em cadeia). Nós descrevemos um método para o isolamento do DNA do vírus do papiloma humano de amostras cervicais utilizando o detergente guanidina (solução DNAzol). O método foi rápido, não-tóxico e eficiente. Uma banda de DNA de 450 pb correspondente à região tardia (L1) do genoma viral foi detectada por PCR, mostrando que a extração com DNAzol gerou quantidade suficiente de DNA para análise. O padrão eletroforético, após digestão com endonucleases de restrição (RFLPs/PCR), revelou predominância de HPV 16 e HPV-33 nas amostras no Estado de Alagoas, Brasil.
HUMAN AND ANIMAL HEALTH
High quality DNA from human papillomavirus (HPV) for PCR/RFLPs
Denise Wanderlei-SilvaI, * * Author for correspondence ; Mariana NobreIII; Rosa Silva GonzagaII; Luciana Silva VianaII; Eduardo Ramalho NetoII
IDepartamento de Patologia; CCBi; Universidade Federal de Alagoas - UFAL; 57010-020; dmws@ofm.com.br; Maceió - AL - Brazil
IILaboratório de Genética Molecular; Genômica e Proteômica - GEMPRO; CECA; Universidade Federal de Alagoas - UFAL; 57100-000; Maceió - AL - Brazil
IIISeção de Patologia Cervical, Hospital Universitário; Universidade Federal de Alagoas - UFAL; 57072-970; Maceió - AL - Brazil
ABSTRACT
The analysis of DNA in clinical samples for a secure diagnostic has become indispensable nowadays. Techniques approaching isolation of high molecular weigth DNA of HPV could lead to efficient amplification and early clinical diagnosis of the virus DNA by PCR (polymerase chain reaction). We describe a fast, non-toxical, efficient and cheap method for DNA isolation of human papilloma virus (HPV) from cervical smears using guanidine (DNAzol solution). A 450 bp DNA band correponding to the late region (L1) of the virus genome was detected by PCR, showing that the DNAzol extraction soluction generated a good viral DNA yield. The electrophoretic pattern after digestion with restriction endonucleases (RFLPs/PCR) revealed the predominance of HPV-16 and HPV-33 in the samples from the State of Alagoas, Brazil.
Key words: Human papillomavirus, DNA purification, PCR, RFLP
RESUMO
A detecção de DNA em amostras clínicas visando um diagnóstico mais seguro vem se tornando uma prática comum em laboratórios de análise clínica. Metodologias que objetivem o isolamento de DNA de alto peso molecular de HPV podem levar a uma amplificação precisa e diagnose precoce do DNA do vírus por PCR (reação de polimerase em cadeia). Nós descrevemos um método para o isolamento do DNA do vírus do papiloma humano de amostras cervicais utilizando o detergente guanidina (solução DNAzol). O método foi rápido, não-tóxico e eficiente. Uma banda de DNA de 450 pb correspondente à região tardia (L1) do genoma viral foi detectada por PCR, mostrando que a extração com DNAzol gerou quantidade suficiente de DNA para análise. O padrão eletroforético, após digestão com endonucleases de restrição (RFLPs/PCR), revelou predominância de HPV 16 e HPV-33 nas amostras no Estado de Alagoas, Brasil.
INTRODUCTION
More than 100 different types of human papillomavirus (HPV) have now been identified by molecular techniques of which almost 30% are mucosotropic (Harnish et al. 1999). They can infect the anogenital tract and contain DNA of high-risk type which is strongly associated with cervical carcinoma (Villa 1997, Walboomers et al. 1999, Wilczynski et al. 1988). Assessment of samples for HPV infection with PCR amplification is important for early detection of initial lesions and virus typing.
The isolation of DNA is an essential step for any technique in Molecular Biology. There have been several recent reports describing different protocols for viral DNA/RNA suitable for PCR amplification (Van der Brule et. al. 1990, Eluf-Neto et al. 1994). The majority of them uses harzadous chemicals, such as phenol and chloroform (Wilczynski et al. 1988), which are laborious, expensive and time consuming. They also involve several steps and are time-consuming. We describe a modification of a protocol using guanidine detergent (DNAzol) for purification of HPV DNA suitable for PCR amplification. The main objective of this work was to develop a fast, high reliable and low cost protocol for isolation of quality woman HPV DNA from cervical samples.
MATERIALS AND METHODS
Sample preparation
A total of 100 cervical smears samples were collected from women attending the Cervical Pathology service of the Federal University of Alagoas Hospital and from private clinics of the State of Alagoas by sampling cells from endocervix and ectocervix with a brush. We selected women with history of any previous suggestive Papanicolaou (Pap), cervical intraepithelial neoplasia (CIN) I, II, and III. The material was placed into Eppendorf tubes containing 500µL of DNA extraction solution (DNAzol Life Technologies) and after 4 to 5 days were sent to the Molecular Genetics Laboratory of the Federal University of Alagoas.
DNA isolation
Genomic DNA isolation was performed by modifying a protocol based on guanidine detergent, the DNAzol solution. The tubes with the samples were inverted several times and DNA precipitation was done by adding 250µL ice-cold isopropanol. The tubes were centrifuged for 1 to 2 min at 4300xg to collect the DNA pellets and the supernatant was carefully discarded. The pellets were washed with 800µL of 95% ethanol by inverting tubes three to six times. The ethanol was removed and tubes were stored for 15 minutes in order to dry the DNA at room temperature. The pellets were ressuspended in 100µL of 8mM NaOH and stored at 4º C.
PCR amplification and restriction analysis
PCR amplification of HPV L1 region was accomplished using primers MY09 (5'-CGTCCMAARGGAWACTGATC-3') and MY11 (5'-GCMCAGGGWCATAAYAATGG-3'). The co-amplification of the human b-globin gene (approximately 268 bp) was performed by using primers GH20 (5'-GAAGAGCCAAGGACAGG TAC-3') and PC04 (5'-CAACTTCATCCACGTT CACC-3'), according to Bauer et al. (1992). Two types of controls were also included in each reaction series: 'no-DNA' and 'HPV-positive DNA'.
All clinical samples employed in this study were positive for b-globin gene after PCR reaction with globin primers. PCR amplification reactions were performed in 100µL, with the following components and final concentrations: 1X Taq buffer, 200µM deoxiribonucleoside triphosphates (dNTPs), 1.5mM MgCl2, 1.0µM of primers MY09/MY11 and 1.0µM of primers GH20/PC04, 2U Taq DNA polimerase, DNA template, 20ng/100µL. PCR reactions were subjected to 35 cycles of amplification on a thermal cycler (Hybaid) using the following program: denaturation at 94ºC for 30 sec, annealing at 55ºC for 1 min, extension at 72ºC for 1 min, and final extension at 72ºC for 8 min. Amplification products were separated by electrophoreses in 1.5% agarose gels, stained with ethidium bromide and photographed under UV light using a Kodak digital system.
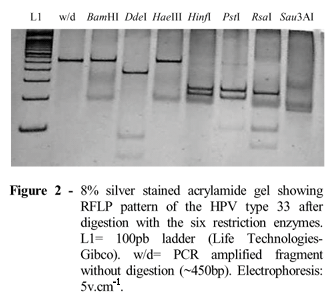
The results of amplification reactions were assessed by scoring sample as either HPV-positive or -negative. Viral PCR products of 450 pb were cleaved with the restriction enzymes: BamHI, Ddel, HaeIII, HinfI, PstI, RsaI, and Sau3AI (Life Technologies), according to Villa (1997). Digested fragments were detected by 8% acrylamide electrophoresis and stained with silver nitrate (Sanguinetti et al., 1994).
RESULTS
DNAzol extraction solution yielded genomic DNA of high molecular weight with fragments of about 23 kb. PCR amplification of HPV late (L1) region accomplished using primers MY09 and MY11 generated fragments of approximately 450 pb (Fig. 1).
Approximately 40% of the samples were HPV positive. Amplimers were separated in 1.5% agarose gel (Fig. 1) by electrophoresis (4V.cm-1) and stained with ethidium bromide. The products of the HPV positive samples were cleaved with the restriction enzymes (RFLPs/PCR) generatig physical maps that could be compared to existing HPV maps, allowing identification of different viral types, and consequently their exclusion or inclusion in high-risk types. The electrophoretic pattern after digestion revealed the predominance of HPV-33 (Fig. 2) and HPV-16 (Fig. 3) in the samples analysed.
DISCUSSION
Molecular studies of HPV have been carried out using different methodologies for viral DNA isolation from clinical samples. Typical protocols use phosphate-buffered saline (PBS) (Eluf-Neto et al. 1994), sodium-dodecyl-sulfate (SDS), proteinase K, and phenol extractions (Wilczynski et al. 1988) or unecessary steps such as freezing followed by heating at high temperatures of cell suspension before PCR (Van der Brule et al. 1990).
Clinical samples from smears might contain blood or endogenous enzymes able to prevent PCR reaction. Furthermore, Taq DNA polimerase can be inhibited by chemicals added to buffers to deactivate infectious agents present in blood and clinical smears (Gravitt and Manos 1992).
Despite the use of traditional protocols, kits for genomic DNA extraction are becoming very popular, mostly because they are ready to use and contain non toxic reagents, such as DNAzol Reagent, a genomic DNA isolation reagent which uses a guanidine-detergent lysing solution for selective precipitation of DNA from cell lysate. It is recommended for purification of DNA from solid and liquid samples from animal, yeast, bacteria and plant sources.
DNAzol extraction solution allowed celular lysis and DNA precipitation in simultaneous steps. An advantage of the solution is to allow samples to be stored for longer periods of time without compromising DNA integrity. It also produced good DNA suitable for HPV detection and identification. The method is simple, fast (20 minutes) and generates DNA of high molecular weight. Another advantage is that no griding is necessary to obtain sufficient DNA.
The efficacy of the protocol was determined by PCR amplifying a fragment corresponding to the L1 region of the virus. The 450 bp fragments indicating the presence of the virus in the samples were readly used for assignment of HPV type by restriction enzyme pattern (RFLP). The physical maps generated allowed identification of different viral types, according to Bernard et al. (1994), and consequently their exclusion or inclusion in high-risk types. Human papilloma virus has been classified in more than 100 types according to its malignat potential assessed by molecular techniques (Harnish et al. 1999). The most common high-risk types HPVs are 16, 18, 31, 33, 35, 51, and 52, which have been associated with CIN II, CIN III and invasive cervical cancer (Eluf-Neto et al. 1994). Our preliminar data show that the predominant types in the State of Alagoas are HPV 16 and HPV 33, both of them presenting high oncogenic potential. Our investigation further support the role of these high-risk types HPVs in the development of cervical neoplasia.
ACKNOWLEDGEMENTS
This work was financially supported by CNPq, Brazilian National Council for Research and Development and FAPEAL, Alagoas State Research Foundation, Brazil.
Received: February 07, 2002;
Revised: September 25, 2003;
Accepted: August 30, 2004.
- Bernard, H. U.; Chan, S. Y.; Manos, M. M.; Ong, C. K.; Villa, L. L.; Delius, H.; Peyton, C. L.; Bauer, H. M. and Wheeler, C. M. (1994), Identification and assessment of known and novel human papillomaviruses by Polymerase Chain Reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J. Infect. Dis., 170, 1077-1085.
- Bauer, H. M.; Greer, C. E. and Manos, M. M. (1992) Detection of genital human papillomavirus infection using PCR. In: Herrington, C. S. and McGee, J. O. D. (Eds.). Diagnostic molecular pathology: a practical approach. Oxford : Oxford University Press. pp. 131-152.
- Eluf-Neto, J.; Booth, M.; Muñoz, N.; Bosch, F. X.; Meijer, C. J. L. M. and Walboomers, J. M. M. (1994), Human papillomavirus and invasive cervical cancer in Brazil. J. Cancer, 69, 114-119.
- Gravitt, P. E. and Manos, M. M. (1992), Polymerase chain reaction - based methods for the detection of human papilomavirus DNA. The Epidemiology of cervical cancer and human Papilomavirus. IARC pp. 121-133.
- Harnish, D. G.; Belland, L. M.; Scheid, E. E. and Rohan, T. E. (1999), Evaluation of human papillomavirus-consensus primers for HPV detection by the polymerase chain reaction. Molecular and Cellular Probes, 13, 9-21.
- Sanguinetti, C. J.; Neto, E. D. and Simpson, A. J. G. (1994), Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques, 16, 915-919.
- Van der Brule, A. J. C.; Meijer, C. J. L. M.; Bakels, V.; Kenemans, P. and Walboomers, J. M. M. (1990), Rapid detection oh human papillomavirus in cervical scrapes by combined general primer-mediated and type-specific polymerase chain reaction. J. Clin. Microbiol, 28, 2739-2743.
- Villa, L. L. (1997), Human Papillomaviruses and Cervical Cancer. In: Advances in Cancer Research Academic Press. pp. 322-341.
- Walboomers, J. M. M.; Jacobs, M. V. and Manos, M. M. et al. (1999), Human papillomavirus is a necessary cause for invasive cervical cancer worldwide. J. Pathol, 189, 12-19.
- Wilczynski, S. P.; Bergen, S., W;alker, J.; Liao, S. Y. and Pearlman, L. F. (1988), Human papillomaviruses and cervical cancer: Analysis of hystopathologic features associated with different viral types. Human Pathology, 19, 697-704.
Publication Dates
-
Publication in this collection
15 Apr 2005 -
Date of issue
Jan 2005
History
-
Accepted
30 Aug 2004 -
Reviewed
25 Sept 2003 -
Received
07 Feb 2002