Abstracts
During an inspection in plastic houses in Sapopema, Paraná, 90% of tomato plants showed leaf abnormalities, probably associated with herbicide toxity. However, virus like symptoms developed in selected hosts after mechanical inoculatation. RT-PCR reactions using primers for an internal region within the movement protein gene of TMV and ToMV resulted in the amplification of a 409 bp cDNA fragment only by TMV primers. Deduced amino acids showed 100% identity when compared to TMV movement protein and 94% with ToMV. The RT-PCR protocol was efficient for quick and conclusive determination of virus species. The virus was purified and a polyclonal antiserum was raised for future surveys in tomato crops of Paraná. The partial genomic sequence obtained for TMV-Sapopema has been deposited under the accession number DQ173945, which is the first partial genomic sequence of an isolate of TMV from Brazil in the GenBank, and the first tomato virus isolate from Paraná to have some of its biological and molecular properties determined.
TMV; P30 protein; plasticulture
Durante uma inspeção em cultivos protegidos de tomate em Sapopema, Paraná, foram observadas anormalidades foliares em 90% das plantas, indicando possivelmente a existência de um problema de fitotoxidade causada por herbicidas. Todavia, os sintomas manifestados nas hospedeiras após os ensaios de inoculação mecânica revelaram que os sintomas estariam relacionados a uma infecção por Tobamovirus. As reações de RT-PCR com oligonucleotídeos específicos para uma região interna da proteína de movimento de dois vírus comuns em tomate, TMV e ToMV, resultaram na amplificação de um fragmento de 409 pares de bases, apenas com os oligonucleotídeos específicos para o TMV. Após o sequenciamento, os aminoácidos deduzidos apresentaram identidade de 100% quando comparados com as seqüências das proteínas de movimento de outros isolados do TMV, e 94% de identidade com seqüências do ToMV. A RT-PCR demonstrou ser um método eficiente para a rápida e conclusiva determinação da espécie viral envolvida na infecção do tomateiro. A seqüência parcial do genoma do isolado de TMV de Sapopema, está depositada no GenBank sob o número de acesso DQ173945, sendo esta a primeira seqüência genômica parcial de um isolado de TMV do Brasil, e conforme o nosso conhecimento, o primeiro isolado de TMV do Paraná a ter algumas de suas propriedades biológicas e moleculares determinadas. Um anti-soro policlonal foi produzido, o que permitirá futuros levantamentos da ocorrência de Tobamovirus nas principais áreas de cultivo de tomate do Paraná.
AGRICULTURE, AGRIBUSINESS AND BIOTECHNOLOGY
Detection and identification of TMV infecting tomato under protected cultivation in Paraná State
Rodrigo Martins da SilvaI; Eliezer Rodrigues de SoutoI,* * Author for correspondence ; Júlio Cézar PedrosoI; Ricardo ArakavaII; Álvaro Manuel Rodrigues AlmeidaIII; Antônio Augusto Lazarini BarbozaI; João Batista VidaI
IDepartamento de Agronomia; Universidade Estadual de Maringá; Av. Colombo, 5790; 87020-900; Maringá - PR - Brasil
IILaboratório de Bioquímica Fitopatológica; Instituto Biológico; Av. Conselheiro Rodrigues Alves, 1252; 04014-002; São Paulo - SP - Brasil
IIIEmbrapa Soja; C. P.: 231; 86001-970; Londrina - PR - Brasil
ABSTRACT
During an inspection in plastic houses in Sapopema, Paraná, 90% of tomato plants showed leaf abnormalities, probably associated with herbicide toxity. However, virus like symptoms developed in selected hosts after mechanical inoculatation. RT-PCR reactions using primers for an internal region within the movement protein gene of TMV and ToMV resulted in the amplification of a 409 bp cDNA fragment only by TMV primers. Deduced amino acids showed 100% identity when compared to TMV movement protein and 94% with ToMV. The RT-PCR protocol was efficient for quick and conclusive determination of virus species. The virus was purified and a polyclonal antiserum was raised for future surveys in tomato crops of Paraná. The partial genomic sequence obtained for TMV-Sapopema has been deposited under the accession number DQ173945, which is the first partial genomic sequence of an isolate of TMV from Brazil in the GenBank, and the first tomato virus isolate from Paraná to have some of its biological and molecular properties determined.
Key words: TMV, P30 protein, plasticulture
RESUMO
Durante uma inspeção em cultivos protegidos de tomate em Sapopema, Paraná, foram observadas anormalidades foliares em 90% das plantas, indicando possivelmente a existência de um problema de fitotoxidade causada por herbicidas. Todavia, os sintomas manifestados nas hospedeiras após os ensaios de inoculação mecânica revelaram que os sintomas estariam relacionados a uma infecção por Tobamovirus. As reações de RT-PCR com oligonucleotídeos específicos para uma região interna da proteína de movimento de dois vírus comuns em tomate, TMV e ToMV, resultaram na amplificação de um fragmento de 409 pares de bases, apenas com os oligonucleotídeos específicos para o TMV. Após o sequenciamento, os aminoácidos deduzidos apresentaram identidade de 100% quando comparados com as seqüências das proteínas de movimento de outros isolados do TMV, e 94% de identidade com seqüências do ToMV. A RT-PCR demonstrou ser um método eficiente para a rápida e conclusiva determinação da espécie viral envolvida na infecção do tomateiro. A seqüência parcial do genoma do isolado de TMV de Sapopema, está depositada no GenBank sob o número de acesso DQ173945, sendo esta a primeira seqüência genômica parcial de um isolado de TMV do Brasil, e conforme o nosso conhecimento, o primeiro isolado de TMV do Paraná a ter algumas de suas propriedades biológicas e moleculares determinadas. Um anti-soro policlonal foi produzido, o que permitirá futuros levantamentos da ocorrência de Tobamovirus nas principais áreas de cultivo de tomate do Paraná.
INTRODUCTION
Crop production based on plasticulture has allowed substantial gains in yields for many horticultural crops. However, this practice may increase the impact of some biotic and abiotic diseases. Among the viruses reported to cause significant losses in tomato (Lycopersicum esculentum Mill.), the commonly found are Cucumovirus, Begomovirus, Luteovirus, Potexvirus, Potyvirus, Tobamovirus, and Tospovirus (Eiras et al., 2002). Fukuda et al., (1981) classified the species of Tobamovirus into two subgroups according to the location of their origin of assembly. The subgroup 1 viruses, Tobacco mosaic virus (TMV), Tomato mosaic virus (ToMV), Pepper mild mottle virus (PMMoV), Tobacco mild green mosaic virus (TMGMV) and Odontoglossum ringspot virus (ORSV) mainly infect solanaceous plants except ORSV, which is a prevalent virus in orchids (Park et al., 1990; Paul, 1975). TMV is the type member of the genus Tobamovirus. Its virion is a rigid rod (18 nm x 300 nm) consisting of about 2130 identical coat protein subunits stacked in a helix around a single strand of positive sense RNA, 6395 nucleotides in length. The genome of TMV encodes at least four proteins (Goelet et al., 1982). The 3' proximal genes encode a 30-kDa cell-to-cell movement protein, also called P30 protein, and a 17.5-kDa coat protein (Meshi et al., 1987, Citovsky et al., 1992). There are reports about Tobamovirus infection in tomato, mainly from São Paulo, Minas Gerais, and Federal District (Resende and Cupertino, 1996). The host range of some isolates include, Nicotiana tabacum L., Petunia sp, tomato and species of Zinnia for TMV, and Tomato for ToMV; (Caner et al., 1990; Bastos et al., 1999; Maritan and Gaspar, 2001; Moreira et al., 2001; Eiras et al., 2002). In Sapopema, tomato plants growing under plastic houses, showing narrowing and leaf distortion in 90% of the plants, suggested a toxity caused by herbicide residues. However, after mechanical inoculation, virus similar symptoms developed in selected hosts. The objective of this work was to identify at species, a tomato virus found in Sapopema, determine some of its biological and molecular properties, and to produce an antiserum for future surveys in tomato producing areas of Paraná State.
MATERIAL AND METHODS
Source and maintenance of the virus
Leaves of tomato 'Santa Clara' showing narrowing, light and dark green mottled areas, producing fruits reduced in size and number with uneven ripening, cultivated in plastic houses in Sapopema-PR, were collected for host range, serology, electron microscopy and RT-PCR analysis. The virus isolate was purified by local lesion transfers on Nicotiana tabacum L. 'Debney', before systemic propagation in 'Santa Clara'. Four hundred grams of leaves were collected from infected plants and cut in 1 cm2 pieces, placed in glass bottles, filled with 300 ml of phosphate buffer (pH 7.0, 0.02M), plus the same volume of glycerol and stored at -20ºC. Fresh material collected from symptomatic plants or the stored material was used for virus propagation in 'Santa Clara' tomato to conduct this work.
Host range
One gram of symptomatic leaves of 'Santa Clara' tissue was ground in 2 ml of 0.01M sodium phosphate buffer (pH 7.0; 1:2 w/v). The extract was rubbed onto leaves of several botanical species, previously dusted with Carborundum® 300 mesh. Symptoms were evaluated at 10 and 20 days after inoculation. The presence of the virus in asymptomatic plants was verified by back inoculation to 'Santa Clara' the same species in which the virus was propagated.
Virus purification
Infected leaves of tomato were harvested approximately 30 days after inoculation. The virus particles were purified by adding 20% of polietileneglicol (PEG) and 5M NaCl according to Gooding and Hebert (1967). Further, the semi-purified preparation was submitted to a linear density sucrose gradient (10-40%), and ultra centrifuged at 80,000g for 2.5h. To determine the yields, dilutions of small aliquots of the preparation were measured at the absorbance of 260 and 280 nm. The yield was estimated assuming an extinction coefficient of three for tobamovirus according to Gibbs and Harrison (1976).
Antiserum production and serological analysis
Antiserum was produced by mixing equal volumes of purified viral solution (containing approximately 5 mg/ml) with Freund's complete adjuvant (Difco) and intramuscular injection of rabbit. Second, third, and fourth injections were prepared with incomplete adjuvant, containing the same amount of virus and made at 10-day intervals. Bleedings began two weeks after the first injection and continued for six weeks. Double diffusion tests were performed according to Purcifull and Bachelor (1977).
Electron microscopy
Small pieces of leaves from infected plants were fixed in a modified Kanovsky fixative (2.5% glutaraldehyde, 2.5% paraformaldehyde in 0.05M cacodylate buffer, pH 7.2), post-fixed with 1% OsO4, dehydrated in acetone and embedded in Spurr's low viscosity resin. Sections were stained with uranyl acetate and Reynold's led citrate and examined in a Zeiss EM 900 transmission electron microscope. Viral particles were also observed in leaf dip preparations negatively stained with uranyl acetate (Kitajima, 1965).
Total nucleic acid extraction and reverse transcription
Total RNA was extracted from 100 mg of fresh leaf tissue ground in liquid nitrogen according to the RNeasy® Plant Kit protocol (Qiagen®, Chatsworth, CA). The cDNA was synthesized using reverse transcriptase (Gibco BRL) and buffers according to the manufacturer in a final volume of 30µl. From the total RNA extraction solution, 10 ml were taken and mixed with 2µl of 10 µM reverse primer TMV-2 (5'-GAA AGC GGA CAGAAA CCC GCT G-3' ) or 2µl of reverse primer ToMV-5 (5'-CTC CAT CGT TCA CAC TCG TTA CT-3') (Jacobi et al, 1998). Afterwards, the samples were incubated at 70 ºC for 10 min and quenched on ice for at least 1 min. The other reaction components were added in the following order: 3µl of 10X PCR buffer (200 mM Tris-HCl, pH 8,4), 500 mM KCl, 2,5 mM MgCl2 (2µl), 10 mM dNTPmix (1 ml), 0.1M dithiothreitol (2µl) and 1µl (20 U) of M-MLV reverse transcriptase (Gibco BRL), and DEPC treated water to give a reaction volume of 30µl. The tubes were incubated for 1h at 42 ºC on an Eppendorf thermocycler model mastercycler personal followed by 10 min of heating at 65 ºC to denature the enzyme.
Amplification by PCR
Reverse primer TMV2 and forward primer TMV1 (5'-GAC CTG ACA AAAATG GAG AAG ATC T-3') are expected to amplify a 422 bp PCR product within the viral movement protein gene of TMV, while primer ToMV-5 and forward primer ToMV-6 (5'-GAT CTG TCA AAG TCT GAG AAA CTT C-3') amplify a product of 508 bp within the same gene of ToMV (Jacobi et al., 1998). PCR reactions were performed in a 40µl volume, containing 10mM each of dATP, dCTP, dGTP and dTTP, 2µl of each primer (10 µM), 2.5 U of Taq DNA Polymerase (Gibco BRL), 4µl of 10X PCR buffer, 25mM MgCl, and 4µl of the first strand synthesis product. Thirty-five reaction cycles were performed as follows: template denaturation at 94 ºC for 30s, primer annealing at 62 ºC for 45s, and elongation at 72ºC for 1 min. A final 5 min elongation step at 72ºC was performed at the end of the 35 cycles.
Sequence comparison
The RT-PCR product was direct sequenced by the dideoxy chain termination method, using a 377 DNA sequencer (ABI, USA). Nucleotide sequence was compared with those available in GenBank (http://www.ncbi.nih.gov/gorf/gorf. html), and the amino acid sequence was estimated through the translation tool from Expasy Software (http://us.expasy.org/tools/). Multiple sequence alignments and sequence relationship were produced using CLUSTAL W (Thompson et al., 1994), and calculation of percentage of identity was performed by analysis on http:// bioinformatics.org/ sms/ ident_sim.html.
RESULTS AND DISCUSSION
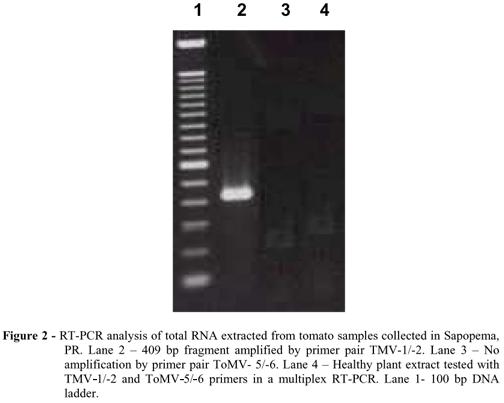
Tomato production of Paraná accounts for approximately 3.4% of the total country production and the north region, where Sapopema is located, concentrates 60% of this (Seab/Deral, 2006). TMV-Sapopema could not infect Petunia hybrida described as a host for TMV and ToMV (Moreira et al., 2003; Hollings and Huttinga, 1976). However, severe mosaic symptoms developed in tomato 'Santa Clara' and 'Avansus' with strong leaf narrowing, while the cultivar Carmem developed only mild symptoms. Misinterpretation of host range symptoms for species determination in Tobamovirus is very common. There are reports of isolates capable to overcome the resistance of some tomato varieties expressing, for example, the resistance factor 'Tm-1' that provides resistance for ToMV, and the resistance factors 'Tm-2' and 'Tm-22' for TMV (Moreira et al., 2003). Additionally few indicator plants distinguish reliably between TMV and ToMV (Jacobi et al. 1998). In this work, electron microscopy examinations of ultratin sections revealed rod shaped particles, and combined with host range, allowed to conclude that a Tobamovirus was associated with the diseased tomatoes from Sapopema (Table 1 and Fig.1). The method for virus purification yielded an average of 5 mg/ml. The antiserum produced reacted to leaf extracts from symptomatic plants in homologous serological reactions and not with healthy plants. According to some authors, serological cross-reaction between ToMV and TMV limits the efficiency of serological differentiation of these two viruses (Jacobi et al., 1998). In this regard, low cross-reaction was observed by Duarte et al., (2002) for a monoclonal antiserum that showed strong specificity for ToMV, but problems for raising this kind of antibodies limited their immediate use. In this work, definitive determination of virus species was possible after RT-PCR assays that reliably discriminated these two viruses. The TMV-1/-2 primer pair was very effective to amplify a 409 bp product located in the TMV movement protein gene (Fig.2).
Sequence analysis of deduced amino acids showed 100% identity with part of the P30 protein of TMV isolates from China and Japan, 94% for ToMV isolates from Australia and Canada and less than 88% with other tobamoviruses (Table 2). This sequence revealed two domains of TMV P30 movement protein responsible for the virus cell-to-cell spread through intercellular connections, as described by Citovsky et al., (1992) (Fig. 3). After many attempts, amplification of this genomic region by primer pair ToMV-5/-6 was not effective, demonstrating the specificity of this primer set for its own virus species (Fig.2). The antiserum produced would facilitate future surveys of TMV and ToMV in combination with RT-PCR, allowing quick and precise species differentiation of this two Tobamoviruses. To our knowledge, this work detailed for the first time, some biological and molecular properties of a TMV isolate from Paraná, responsible for production decrease in two plasticulture locations in Sapopema.
ACKNOWLEDGMENTS
We are grateful to Dr. Elliot Watanabe Kitajima (NAPA-USP-Esalq) for his contribution to the electron microscopy work.
Received: August 29, 2005;
Revised: October 22, 2007;
Accepted: June 20, 2008.
- Bastos, H.B., Pavan, M.A. and Silva, N. (1999), Estirpes do vírus do mosaico do tomateiro presentes em regiões produtoras de tomate do Estado de São Paulo. Summa Phytopathologica 1,14-16.
- Caner, J., Colariccio, A., Chagas, C.M., Alba, A.P.C. and Vicente, M. (1990), Identificação de um isolado do vírus do mosaico do tomateiro (ToMV) no Estado de São Paulo. Fitopatologia Brasileira 15, 347-350.
- Citovsky, V., Wong, M. L., Shaw, A. L, Prasad, B., V.,V. and Zambryski, P. (1992), Visualization and characterization of Tobacco Mosaic Virus movement protein binding to single-stranded nuclei acids. Plant Cell 4, 397-411.
- Duarte, K.M.R., Gomes, L.H., Andrino, F.G., Leal Jr., G.A., Silva, F.H.B., Paschoal, J.A.R., GiacomelliI, A.M.B. and Tavares, F.C.A. (2002), Identificação do vírus do mosaico do tomateiro (ToMV) Tobamovirus, por meio de anticorpos monoclonais. Scientia Agrícola 59, 107-112
- Eiras, M., Chaves, A.L.R., Colariccio, A., Harakava, R., Araujo, J. and Chagas, C.M. (2002), Caracterização do Tomato chlorotic spot tospovirus isolado de jiló no Estado de São Paulo. Fitopatologia Brasileira 27, 285-291.
- Fukuda, M., Meshi, T., Okada, Y., Takebe, I. (1981), Correlation between particle multiplicity and location on virion RNA of the assembly initiation site for viruses of the tobacco mosaic virus group. Proceedings of the National Academy of Science USA 78, 4231-/4235.
- Goelet, P., Lomonossoff, G.P., Butler, P., J., G., Akam, M.E., Gait, M.J, Karn, J. (1982), Nucleotide sequence of tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 79, 581822.
- Gooding, J.V., Hebert, T.T. (1967), A simple technique of purification of tobacco mosaic virus in large quantities. Phytopathology 57, 1285.
- Gibbs, A. and Harrison, B.D. (1976), Plant virology. The principles. Edward Arnold Publication. London.
- Hollings, M. and Huttinga, H. (1976), Tomato mosaic virus. CMI/ AAB. Descriptions of plant viruses. nş. 156.
- Jacobi, V., Bachand, G.D., Hamelin, R.C., Castello, J.D. (1998), Development of a multiplex immunocapture RT-PCR assay for detection and differentiation of tomato and tobacco mosaic tobamoviruses. Journal of Virological Methods 74, 167-178.
- Kitajima, E.W. (1965), Electron microscopy of vira cabeça virus within the host cell. Virology 26, 89-99.
- Maritan, A.C. and Gaspar, J.O. (2001), Infecção de Zinnia elegans pelo Tobacco mosaic virus resultante da interação com um potyvírus. Fitopatologia Brasileira 26, 533.(Resumo).
- Meshi T, Watanabe Y, Saito T, Sugimoto A, Maeda T, Okada Y. (1987), Function of the 30-kd protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J. 6, 255763
- Moreira, S.R., Colariccio, A., Eiras, M. and Chaves, A.L.R. (2001), Caracterização molecular do Tomato mosaic virus isolado de tomateiro no Estado de São Paulo. Fitopatologia Brasileira 26, 515. 2001 (Resumo).
- Moreira, S.R., Eiras, M., Chaves, A.L.R., Galleti, S.R. and Colariccio, A. (2003), Caracterização de uma nova estirpe do Tomato mosaic virus isolada de tomateiro no Estado de São Paulo. Fitopatologia Brasileira 28, 602-607.
- Park, W.M., Kim, W.G., Ryu, K.H., Yoon, K.E., Kwak, B.H., So, I.S. (1990), Detection of odontoglossum ringspot virus and cymbidium mosaic virus from cultivated orchids by immunosorbent electron microscopy. Journal of the Korean Society of Horticultural Sciences 31, 417-422.
- Paul, H.L. (1975). Odontoglossum ringspot virus. CMI/AAB. Description of Plant Viruses No. 155, Kew, England.
- Purcifull, D.E. and Batchelor, D.L. (1977). Immunodiffusion tests with sodium dodecyl sulphate (SDS)-treated plant virus and plant virus inclusions. Gainesville: University of Florida. (Bulletin, 788). 39p.
- Resende, R.O. and Cupertino, F.P. (1996). Doenças causadas por vírus em tomateiro. Informe Agropecuário, Belo Horizonte, 18, n.184. p.19-27.
- SEAB/DERAL. (2006). Valor bruto da agropecuária paranaense. Disponível em: http:/www.pr.gov.br/seab/deral Acesso em: 4 out. 2006.
- Thompson, J.D., Higgins, D.G. and Gibson, T.J. (1994). Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position, specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 467-4680.
Publication Dates
-
Publication in this collection
29 Oct 2008 -
Date of issue
Oct 2008
History
-
Accepted
20 June 2008 -
Reviewed
22 Oct 2007 -
Received
29 Aug 2005