Abstracts
In this study, the abundance of IGF-II and bFGF transcripts was estimated in the chicken embryos using the competitive RT-PCR analysis. Significant enhancements in the abundance of IGF-II mRNA were observed at stages HH1 and 5, and a new accumulation in these levels was observed at stage HH18 in comparison to the basal levels. The abundance of bFGF mRNA increased significantly at stages HH18 and 20, followed by an upregulation in the expression of these transcripts at stage HH26. These findings provided important information about the temporal expression pattern of IGF-II and bFGF transcripts in the whole chicken embryos during in ovo development.
chicken embryos; temporal expression; competitive RT-PCR; growth factors
Fatores de crescimento coordenam múltiplas vias de sinalização durante o desenvolvimento embrionário. Neste estudo, a abundância de mRNA dos genes IGF-II e bFGF foi estimada em embriões de galinha por análises de RT-PCR competitiva. Aumentos na abundância de mRNA de IGF-II foram observados nos estádios HH1, 5. Os níveis de mRNA de bFGF exibiram aumentos a partir dos estádios HH18 e 20, seguido por uma acentuada redução a níveis basais no estádio HH24 e por um segundo pico na expressão destes transcritos no estádio HH26. Tais descobertas proporcionam importantes informações sobre o padrão de expressão destes fatores de crescimento durante a embriogênese de aves
HUMAN AND ANIMAL HEALTH
Temporal expression pattern of the insulin-like growth factor II and fibroblast growth factor transcripts in avian embryogenesis
Jane Eyre GabrielI,* * Author for correspondence ; Lúcia Elvira AlvaresII; Luiz Lehmann CoutinhoIII
ICentro de Ciências Exatas e Sociais Aplicada; Universidade Estadual da Paraíba; 58700-08; Patos - PB - Brasil
IIInstituto de Biologia; Universidade Estadual de Campinas; 13083-970; Campinas - SP Brasil
IIIDepartamento de Produção Animal; Escola Superior de Agricultura Luiz de Queiroz; Universidade de São Paulo; 13418-900; Piracicaba - SP - Brasil
ABSTRACT
In this study, the abundance of IGFII and bFGF transcripts was estimated in the chicken embryos using the competitive RT-PCR analysis. Significant enhancements in the abundance of IGF-II mRNA were observed at stages HH1 and 5, and a new accumulation in these levels was observed at stage HH18 in comparison to the basal levels. The abundance of bFGF mRNA increased significantly at stages HH18 and 20, followed by an upregulation in the expression of these transcripts at stage HH26. These findings provided important information about the temporal expression pattern of IGF-II and bFGF transcripts in the whole chicken embryos during in ovo development.
Key words: chicken embryos, temporal expression, competitive RT-PCR, growth factors
RESUMO
Fatores de crescimento coordenam múltiplas vias de sinalização durante o desenvolvimento embrionário. Neste estudo, a abundância de mRNA dos genes IGF-II e bFGF foi estimada em embriões de galinha por análises de RT-PCR competitiva. Aumentos na abundância de mRNA de IGF-II foram observados nos estádios HH1, 5. Os níveis de mRNA de bFGF exibiram aumentos a partir dos estádios HH18 e 20, seguido por uma acentuada redução a níveis basais no estádio HH24 e por um segundo pico na expressão destes transcritos no estádio HH26. Tais descobertas proporcionam importantes informações sobre o padrão de expressão destes fatores de crescimento durante a embriogênese de aves.
INTRODUCTION
The animal embryogenesis is remarkably equipped for precise mechanisms of the cellular proliferation and differentiation until the formation of functional tissue. Insulin-like growth factors (IGFs) and fibroblast growth factors (FGFs) are mitogenic agents that play fundamental regulatory roles during the embryo development, since these peptides regulate cell fate by activating the multiple transduction pathways.
Somatomedins (IGF-I and II) comprise a family of the polypeptides with the growth-promoting effects in a wide variety of the cultured cells (Florini et al., 1991; Florini et al., 1996; Pirskanen et al., 2000). It has been shown that these factors stimulate the proliferation, differentiation and protein accretion of the cells derived from the avian embryos (Szebenyi and Fallon, 1999). In vitro analysis shows that IGFs promote the terminal cellular differentiation for an expression- and secretion-dependent autocrine mechanism. Contrary to that observed in the mammals, IGF-I and -II exert similar effects in the chicken embryos due to the absence of type-II IGF receptors. The IGF-II gene is essential for the normal embryonic and postnatal growth and plays an important role in the functioning of a healthy immune system, lymphopoiesis, myogenesis and bone growth among other physiological functions (Denley et al., 2005).
The members of the FGF family are important molecules involved at the early steps of the morphogenetic processes and include the major regulators of the embryonic development (Sanchez et al., 2001). They influence the formation of the primary body axis, neural axis, limbs and other embryonic structures. The activities of the FGFs depend on their coordination of the fundamental cellular functions, such as survival, replication, differentiation, adhesion and motility, through the effects on gene expression and on the cytoskeleton (Szebenyi and Fallon, 1999). The basic fibroblast growth factor, also known as bFGF or FGF2, is a particularly potent mitogen and promotes the cell survival by inhibiting the apoptosis. At the tissue level, it is involved in wound repair and induces the angiogenesis (Muramatsu et al., 2002). Additionally, bFGF is a significant target in the cancer research because it is over-expressed in some cancers and may enhance a tumor's metastatic potential (Castelli et al., 2004).
Although the effects of the growth factors on the cultured cells have been extensively reported (Florini et al., 1996), their expression pattern and relationship with the development processes of the embryogenesis has not been well developed. Thus, the aim of this study was to estimate the abundance of the IGF-II and bFGF mRNAs in chicken embryos throughout different developmental stages in order to verify the temporal expression pattern of these growth regulatory genes during in ovo development.
MATERIAL AND METHODS
Embryo collection and total RNA extraction
Fertilized chicken eggs from Cobb500® line were incubated at 37±0.5º C and 60% relative humidity. Fifteen embryos were surgically collected and staged according to Hamburger and Hamilton (1951). The analyzed developmental stages were HH1 (blastoderm stage, unincubated embryo), 5, 8, 10, 12, 15, 18, 20, 21, 23, 24 and 26 (fifth day of incubation). After the removal of the membranes, the whole embryos were immediately dissolved in TRIZOL reagent (Invitrogen). Total RNA extraction was performed as described by Chomczynski and Sacchi (1987). These samples were resuspended in 20-100 µl of sterile nuclease-free water for the determination of their concentrations. The integrity of the ribosomal RNA bands was observed on agarose gels under the denaturant conditions (Sambrook et al., 1989).
RT-PCR analysis
Single-strand cDNA synthesis was carried out from 1 µg of the total RNA by the reverse transcription (RT) reaction. After the denaturation at 70º C for 10 min, RNA samples were incubated in 1X PCR buffer (10 mM Tris-HCl, pH 9.5, 50 mM KCl, 0.1% Triton X-100), 2.5 mM MgCl2, 1 mM dNTPs mix, 500 ng oligo(dT) primer, and 200 U SuperScriptII enzyme (Invitrogen) in a final volume of 20 µl. This reaction was maintained at 42º C for 50 min and subsequently incubated with 1 U of RNase H for 20 min at 37º C for the RNA template digestion.
The amplification reactions were conducted using the sense and antisense primers as shown in Table 1. All the primers were designed to flank an intron to avoid the same-size amplification of the contaminating genomic DNA. The PCRs were performed with 10% of total RT reaction volume, 0.5 µM specific primers (Table 1), 1X PCR buffer (10 mM Tris-HCl, 1.5-2.5 mM MgCl2, 50 mM KCl), and 1 U Taq DNA polymerase (Invitrogen). These reactions were denatured at 94º C for 30 seconds, followed by annealing for 40 seconds and extension corresponding to 72º C for 40 seconds. The annealing temperatures for IGF-II and bFGF genes were 57 and 58º C, respectively and the cycle number ranged from 20 to 40 in function of the mRNA abundance. The identity of the amplified products was confirmed by digestion with the restriction enzymes (Table 1).
Construction of native and competitor fragments
The mRNA expression levels of IGF-II, bFGF and β-actin genes were estimated using a modified experimental procedure of the quantitative-competitive RT-PCR initially described by Tsai and Wiltbank (1996) and reported in detail by Gabriel et al. (2003). Two cDNA fragments for each gene were generated in order to construct the standard curves. To synthesize the native cDNA fragments of the genes of the interest, the RT reaction was performed exactly as described above, since these fragments were identical in size and nucleotide sequence to the cDNA obtained by the RT-PCR of the embryos. The competitor cDNA fragments were generated using the identical primer binding sites with the native template and these fragments were produced by mutational PCR with the antisense deletion primers (Table 1). The deletion primers were composed of 19 bases complementary to an internal region of the target template in addition to 20 bases identical to the antisense primer. After the amplification, the region downstream from the deletion primer site was eliminated in the competitor, resulting in a smaller fragment. However, the deletion primer contained the complete sequence of the antisense primer site and this stretch was rescued in the copied fragment. Consequently, the same primer pairs could be employed to amplify both the native and competitor cDNAs. Since the region to which primers bind was common, the competitor and native DNA fragments competed equally for the limiting reaction components (nucleotides, primers and enzyme).
The native and competitor fragments were amplified from 10% of RT reactions, eluted from the agarose gel using the Sephaglass Elution kit (Amersham Pharmacia Biotech) and cloned in the expression vectors using the SureClone Ligation kit (Amersham Pharmacia Biotech). To confirm the identity and the deletion of the desired sequences, these clones were sequenced with the BigDye Terminator kit (Applied Biosystems) using an ABI PRISM 377 automated DNA sequencer.
Construction of standard curves and determination of expression levels of interest genes
The transcript abundance was estimated by the direct comparison with the standard curves obtained from increasing and known amounts of the native cDNA in the presence of the constant amount of the competitor cDNA (Gabriel et al., 2003). Serial dilutions of the native cDNA fragment were performed in the water containing 10 ng/µl of the plasmidial DNA (with no insert) as the carrier. The amount of the native cDNA added to each PCR varied from 25.6 X 10-n to 0.1 X 10-n while a fixed amount of 1 X 10-n of the competitor fragment was maintained, where n corresponded to co-amplification range in which each standard-curve was constructed.
The amount of the competitor fragment used for each gene was determined by the titration experiments, in which a different amount of the competitor cDNA was co-amplified with the experimental sample. The co-amplification range for IGF-II and bFGF genes was 10-5. The β-actin expression levels were determined in order to correct the variations in the total RNA quantification and in reverse transcription efficiency, and thus, normalize the mRNA abundance of genes of interest.
A known amount of the competitor was directly added to the 1 X PCR master mixture to assure that an equal amount was present in all the samples analyzed. To monitor the possible DNA contaminations, negative controls were included in each batch of the PCR. Sense primers were 5'-labelled with the fluorescein and aliquots of the amplified products were submitted to the electrophoresis in an Automated Laser Fluorescent DNA Sequencer (A.L.F.TM, Pharmacia LKB Biotechnology). Following the electrophoresis, the peaks derived from the native and competitor templates were integrated with the Fragment Manager software (Pharmacia LKB Biotechnology). The logarithmic proportion of the native to competitor cDNA peak areas (log N/C) and the initial concentration logarithm of the native cDNA (log Ni) were employed for the standard curve construction. Finally, the mathematical equation obtained by the linear regression of the standard curves was used to determine variations in the mRNA abundance of genes of interest in the embryonic cDNA samples based on the ratio of the native to competitor products in individual PCRs.
Statistical analysis
The data obtained in the quantitative assays were statistically analyzed by the regression analysis using the GLM procedure of SAS® (1999). The data corresponding to bFGF gene were submitted to the logarithmic transformation, whereas results concerning IGF-II gene were estimated by the square root transformation as proposed by Cook and Weisberg (1994). The statements of significance were based on P<0.05 unless otherwise noted.
RESULTS AND DISCUSSION
The abundance of IGF-II and bFGF transcripts was estimated in the chicken embryos at different developmental stages using competitive RT-PCR analysis (Fig 1). Three-fold accumulation in IGF-II transcript levels was detected significantly at stages HH1 (blastoderm) and 5 (p<0.05) (Fig 1A).
This could be explained by the fact that after the hatching, the blastoderm presented thousand of the cells in an active process of the cellular division that mobilized all the information needed by the functional developing chicken embryos (Hamburger and Hamilton, 1951). A second significant peak in these levels was detected in the chicken embryos at stage HH18 and 20 in comparison to the basal levels (p<0.05) (Fig 1A).
Thus, the enhancements in the levels of IGF-II mRNA found in the chicken embryos at early developmental stages could indicate its fundamental participation as a potent mitogenic agent, regulating the important pathways of the cellular proliferation during the embryogenesis.
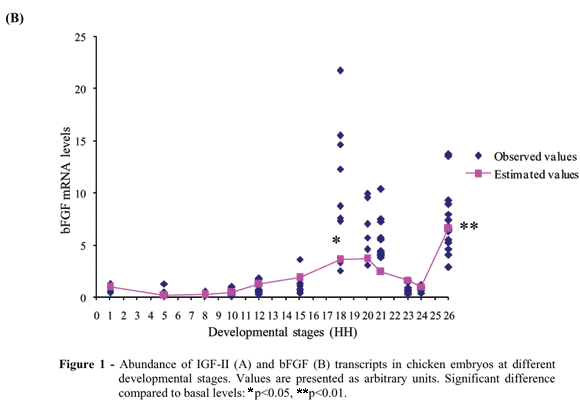
The basal levels in the expression of the bFGF gene were detected from stages HH1 until 15 (p>0.05) (Fig 1B). Significant three-fold enhancements in these levels were found at stages HH18 and 20 (p<0.05) compared to the basal levels (Fig 1B). Although Hannon et al. (1992) found that this factor was not expressed during the early mouse embryonic development, the findings presented in this study showed enhancements in the levels of bFGF mRNA from stage 18 and 20 in the whole chicken embryos (p<0.05) (Fig 1B).
This accentuated expression could trigger the cellular differentiation events and the formation of the bud limb in the chicken embryos, as also observed in the chicken embryos at the same developmental stage by Cohn et al. (1995) and Ohuchi et al. (1997).
A new significant six-fold peak in abundance of bFGF mRNA was observed for stage HH26 compared to the basal levels (p<0.01) (Fig 1B). The upregulation of bFGF mRNA at stage HH26 in the whole chicken embryos reinforced the findings described by Kocamis and Killefer (2003), which observed an accentuated expression of these transcripts in the skeletal muscle tissue isolated from the chicken embryos at the same developmental stage. The evidence presented in this study suggested that the bFGF gene exerted a crucial role in the modulation of the cellular differentiation events in the avian embryogenesis.
Under these experimental conditions, the differences in the transcript abundance between the both growth factors were detected in the embryos at stages 1, 5, 18, 20 and 26 (p>0.01) (Fig 1). Except at stage 26, the levels of IGF-II mRNA were higher than those observed in bFGF (Fig 1). The growth factors act as signaling molecules in a wide variety of biological events in the vertebrates and several studies have focused on the effects that these peptides have on the molecular basis involved in the deposition of the skeletal muscle tissue (Pownall and Emerson, 1992, Bass et al., 1999). Considering that the FGF signaling pathways were intricately intertwined with IGFs (Szebenyi and Fallon, 1999), it could be speculated that these genes could interact synergistically in order to modulate distinct myogenic events during early avian embryogenesis. Further research is necessary to address the complex modulation network of the regulatory gene machinery that these growth factors exert in ovo development.
In conclusion, the findings described in this study provided important insights into the temporal expression pattern of IGF-II and bFGF genes during early avian embryogenesis.
ACKNOWLEDGEMENTS
Authors are grateful to the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for the financial support.
Received: April 27, 2006;
Revised: July 13, 2007;
Accepted: May 06, 2008.
- Bass, J., Oldham, J., Sharma, M., Kambadur, R. (1999), Growth factors controlling muscle development. Dom. An. Endocrinol, 17, 191-197.
- Borja, A.J.M., Zeller, R., Meyers, C. (1993), Expression of alternatively spliced bFGF coding exons and antisense mRNAs during chicken embryogenesis. Dev. Biol., 157, 110-118.
- Castelli, R., Porro, F., Tarsia P. (2004), The heparins and cancer: review of clinical trials and biological properties. Vas. Med., 9, 205-213.
- Chomczynski, P., Sacchi, N. (1987), Single step method of RNA isolation by acid guanidinium thiocyanate-phenol chloroform extraction. Analytical Biochem., 162, 156-159.
- Cohn, M.J., Izpisúa-Belmonte, J.C., Abud, H., Heath, J.K., Tickle, C. (1995), Fibroblast growth factors induce additional limb development from the flank of chick embryos. Cell, 80, 739-746.
- Cook, R.D., Weisberg, S. (1994), Transforming a response variable for linearity. Biometrika, 81, 731-737.
- Darling, D.C., Brickell, P.M. (1996), Nucleotide sequence and genomic structure of the chicken insulin-like growth factor-II (IGF-II) coding region. Gen. Comp. Endocrinol, 102, 283-287.
- Denley, A., Cosgrove, L., Booker, G., Wallace, J., Forbes, B. (2005), Molecular interactions of the IGF system. Cytokine Growth Factor Rev., 16, 421-439
- Florini, J.R., Magri, K.A., Ewton, D.Z., James, P.L., Grindstaff, K., Rotwein, P. (1991), "Spontaneous" differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J. Biol. Chem., 266, 15917-15923.
- Florini, J.R., Ewton, D.Z., Cooligan, S.A. (1996), Growth hormone and the insulin-like growth factor system in myogenesis. Endocrine Rev, 1795, 481-517.
- Gabriel, J.E., Javiel, H.A., Alvares, L.A., Schmidt, G., Coutinho, L.L. (2000). In situ detection of the myogenic factor MyoD in whole chicken embryos. Genet. Mol. Biol, 23, 145-148.
- Gabriel, J.E., Alvares, L.E., Gobet, M.C., de Paz, C.C.P., Packer, I.U., Macari, M., Coutinho, L.L. (2003), Expression of MyoD, myogenin, myostatin and Hsp70 transcripts in chicken embryos submitted to mild cold or heat. J. Thermal Biol., 28, 261-269.
- Hamburger, V., Hamilton, H.L. (1951), A series of normal stages in the development of the chick embryo. J. Morphol., 88, 49-92.
- Hannon, K., Smith, C.K., Bales, K.R., Santerre, R.F. (1992), Temporal and quantitative analysis of myogenic regulatory and growth factor gene expression in the developing mouse embryo. Dev. Biol, 151, 137-144.
- Kocamis, H., Killefer, J. (2003), Expression profiles of IGF-I, IGF-II, bFGF and TGF-b2 growth factors during chicken embryonic development. Turk J Vet Anim Sci., 27, 367-372.
- Kost, T.A., Theodorakis, N., Hughes, S.H. (1983), The nucleotide sequence of the chick cytoplasmic beta-actin gene. Nucleic Acids Res, 11, 8287-8301.
- Muramatsu, M., Yamada, M., Takai, S., Miyazaki, M. (2002), Suppression of basic fibroblast growth factor-induced angiogenesis by a specific chymase inhibitor, BCEAB, through the chymase-angiotensin-dependent pathway in hamster sponge granulomas. Br. J. Pharmacol., 137, 554-560
- Ohuchi, H., Nakagawa, T., Yamamoto, A., Araga, A., Ohata, T., Isbimaru, Y., Yoshioka, H., Kuwana, T., Nobno, T., Yamasaki, M., Itoh, N., Noji, S. (1997), The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development, 113, 1419-1434.
- Pirskanen, A., Kiefer, J.C., Hauschka, S.D. (2000), IGFs, insulin, Shh, bFGF, and TGF-beta 1 interact synergistically to promote somite myogenesis in vitro. Dev. Biol, 224, 189-203.
- Pownall, M.E., Emerson Jr., C.J. (1992), Sequential activation of three myogenic regulatory genes during somite mophogenesis in quail embryos. Dev. Biol, 151, 67-79.
- Sambrook, J., Fritsch, E.F., Maniatis, T. Extraction, purification, analysis of messenger RNA from eukaryotic cells. In: Ford, N. Molecular cloning: a laboratory manual. 2.ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 1989, pp.7.40-7.87.
- Sanchez, C.L., Rodriguez-Gallardo, L., Alvarez, I.S., Climent, V., Garcia-Martinez, V. (2001), Effects of growth factors on the commitment of chick blastoderm. Int. J. Dev. Biol, 45, S109-S110.
- SAS Institute. SAS/STAT User's guide. Online Version. 8.ed. Cary: SAS Institute, 1999.
- Szebenyi G, Fallon JF. (1999), Fibroblast growth factors as multifunctional signaling factors. Int Rev Citol., 185, 45-106.
- Tsai, S., Wiltbank, M.C. (1996), Quantification of mRNA using competitive RT-PCR with standard curve methodology. Biotechniques, 21, 862-866.
Publication Dates
-
Publication in this collection
29 Oct 2008 -
Date of issue
Oct 2008
History
-
Accepted
06 May 2008 -
Reviewed
13 July 2007 -
Received
17 Apr 2006