Abstract
The aim of the present study was to investigate the antimicrobial effect of the lipopeptide biosurfactants produced by Bacillus licheniformis strain M104 grown on whey. The biosurfactant was investigated for potential antimicrobial activity by using the disc-diffusion method against different Gram-positive bacteria {B subtilis, B. thuringiensis (two strains), B. cereus, Staphylococcus aureus (two strains) and Listeria monocytogenes)}, Gram-negative bacteria {(Pseudomonas aeruginosa, Escherichia coli (two strains), Salmonella typhimurium, Proteous vulgaris and Klebsiella pneumoniae) and a yeast (Candida albicans)}. The biosurfactant showed profoundly distinct antibacterial activity toward tested bacteria and displayed an antifungal activity against the tested yeast. Maximum antimicrobial activity of the biosurfactant was shown against S. aureus ATCC 25928. The biosurfactant had a broad inhibition effect on intracellular components of S. aureus ATCC 25928. The antimicrobial effect of lipopeptide biosurfactant produced by B. licheniformis strain M104 was time and concentration dependent. When biosurfactant was added to S. aureus medium in a concentration of (48 μg / ml), the maximum reduction of acid soluble phosphorous (53.06 %), total lipid (90.47 %) total proteins (53.43%), RNA (83.29 %) and DNA (48.50%) were recorded after 12 h of incubation period. From the preliminary characterization results, it could be concluded that biosurfactants were a suitable alternative in potential applications of medical fields.
Biosurfactant; Bacillus licheniformis; Cheese whey; antimicrobial activity; Staphylococcus aureus
BIOLOGICAL AND APPLIED SCIENCES
Antimicrobial activity of a biosurfactant produced by Bacillus licheniformis strain M104 grown on whey
Eman Zakaria Gomaa** Author for correspondence: emann7778@yahoo.com
Department of Biological and Geological Sciences, Faculty of Education, Ain Shams University, Roxy, 11757, Cairo - Egypt
ABSTRACT
The aim of the present study was to investigate the antimicrobial effect of the lipopeptide biosurfactants produced by Bacillus licheniformis strain M104 grown on whey. The biosurfactant was investigated for potential antimicrobial activity by using the disc-diffusion method against different Gram-positive bacteria {B subtilis, B. thuringiensis (two strains), B. cereus, Staphylococcus aureus (two strains) and Listeria monocytogenes)}, Gram-negative bacteria {(Pseudomonas aeruginosa, Escherichia coli (two strains), Salmonella typhimurium, Proteous vulgaris and Klebsiella pneumoniae) and a yeast (Candida albicans)}. The biosurfactant showed profoundly distinct antibacterial activity toward tested bacteria and displayed an antifungal activity against the tested yeast. Maximum antimicrobial activity of the biosurfactant was shown against S. aureus ATCC 25928. The biosurfactant had a broad inhibition effect on intracellular components of S. aureus ATCC 25928. The antimicrobial effect of lipopeptide biosurfactant produced by B. licheniformis strain M104 was time and concentration dependent. When biosurfactant was added to S. aureus medium in a concentration of (48 μg / ml), the maximum reduction of acid soluble phosphorous (53.06 %), total lipid (90.47 %) total proteins (53.43%), RNA (83.29 %) and DNA (48.50%) were recorded after 12 h of incubation period. From the preliminary characterization results, it could be concluded that biosurfactants were a suitable alternative in potential applications of medical fields.
Key words:Biosurfactant, Bacillus licheniformis, Cheese whey, antimicrobial activity, Staphylococcus aureus
INTRODUCTION
Biosurfactants are amphiphilic compounds produced by the microorganisms with pronounced surface and emulsifying activities (Singh et al. 2007). Microbial surfactants comprise a diverse group of surface-active molecules, which are categorized by their chemical composition and microbial origin. They include glycolipids, lipopeptides, polysaccharideprotein complexes, protein-like substances, lipopolysaccharides, phospholipids, fatty acids and neutral lipids (Van Hamme et al. 2006). Therefore, it is reasonable to expect diverse properties and physiological functions of biosurfactants such as increasing the surface area and bioavailability of hydrophobic water-insoluble substrates, heavy metal binding, bacterial pathogenesis, quorum sensing, and biofilm formation (Singh and Cameotra 2004). These compounds can be synthesized by the microorganisms growing on water-immiscible hydrocarbons as well as on water-soluble compounds (Mukherjee et al. 2006).
The antimicrobial activity of several biosurfactants has been reported in the literature for many different applications (Cameotra and Makkar 2004). Some biosurfactants are known to have therapeutic applications as antibiotics and antifungal or antiviral compounds. Among the many classes of biosurfactants, lipopeptides represent a class of microbial surfactant with remarkable biological activities, such as antimicrobial, antitumor, antiviral and antiadhesive activities (Khire and Khan 1994; Banat 1995; Peypoux et al. 1999). These properties make them relevant molecules for applications in combating many diseases and as therapeutic agents.
However, the main factor that restricts the widespread use of biosurfactants is their production cost when compared to their synthetic counterparts (Mukherjee et al. 2006). The cost can be reduced by strain improvement, optimizing the medium composition by statistical methods or by using alternative inexpensive substrates. The choice of inexpensive raw materials is important to the overall economy of the process as they account for 50% of the final production cost and also reduce the expenses with waste treatment (Makkar and Cameotra 1999).
Whey is a liquid by-product of cheese production containing the water-soluble components. It is composed of high levels of lactose (75% of dry matter) and 1214% protein. In addition, organic acids, minerals, and vitamins are present. Whey disposal represents a major pollution problem, especially for the countries depending on dairy economy. Only half of the cheese whey produced annually is recycled into useful products such as food ingredients and animal feed and the rest is regarded as a pollutant (Joshi et al. 2008). These studies showed that whey wastes might be comparatively better substrates for biosurfactant production at the commercial scale and efficient dairy wastewater management.
The aim of the present study was to investigate the antimicrobial effect of the biosurfactant produced by Bacillus licheniformis strain M104 grown on whey. Moreover, the effect of the produced biosurfactant on intracellular components of a pathogenic bacterial strain, Staphylococcus aureus ATCC 25928 was studied.
MATERIALS AND METHODS
Isolation and screening of biosurfactant- producing bacteria
Four soil samples, collected from the oil-contaminated sites of western desert -Egypt, were screened for biosurfactant-producing bacteria by using the modified procedure described by Bodour et al. (2003). Five grams of each soil sample was placed into a 250 ml flask containing 50 ml of tap water and incubated at 30°C ± 2°C on a shaker at 150 rpm for 21 days. On days 3, 7, 14, and 21, a sample from each soil slurry was serially diluted, plated on PYG agar and incubated for three days. After incubation, morphologically different bacteria were selected for biosurfactant screening (approximately 10 to 15 isolates per sampling time). Developed colonies on the plates were then repeatedly sub-cultured on PYG agar medium to obtain pure isolates, and then maintained on the slants of the same medium.
Isolated colonies were inoculated into 50 ml minimal salt medium (MSM) containing 0.5% (v/v) crude oil as the sole carbon and energy source. The broth cultures were incubated with shaking (150 rpm) at 30°C± 2°C for seven days. The cell suspensions were then tested for the presence of biosurfactant by using the qualitative drop-collapse method (Youssef et al., 2004). Briefly, the drop-collapse technique was carried out by adding 100 µl culture supernatant to the wells of a 96-well microliter plate lid, then 5 µl of crude oil was added to the surface of the culture supernatant. Biosurfactant-producing culture gave flat drops. Aliquots from a culture of each strain were analyzed on two separate plates.
Production media and culture conditions
The seed culture was prepared by transferring a loopful from a fresh culture of B. licheniformis strain M104 into 50 ml of peptone yeast glucose medium (PYG) containing (g/l): Peptone, 5; yeast extract, 5; glucose, 15 (Rocha et al., 1992). The flasks were incubated with shaking at 150 rpm at 30oC± 2°C for 48 h.
Erlenmeyer flasks (500 ml volume) containing 100 ml of the modified minimal salt medium of Deziel et al. (1996) modified by Ramadan et al. (2011) was used. It contained the following constituents (g/l): MgSO4.7H2O, 0.2; CaCl2.2H2O, 0.02; KH2PO4, 3; K2HPO4, 3; urea, 2 and FeCl3, 0.05, whey was added as the sole carbon source in a concentration of 10 g/l. Initial pH was adjusted to 7.0 and then inoculated with 10 % of the seed culture (Sepahy et al., 2005). The inoculated flasks were incubated on a rotary shaker (150 rpm) at 30oC±2°C for seven days.
Extraction of biosurfactant
The preparation of biosurfactant was described by Cao et al. (2007). Briefly, the bacterial cells were removed by centrifugation at 10,000 rpm at 4ºC for 10 min. The cell-free supernatant was adjusted to pH 2.0 using 6 N HCl and kept at 4oC for 24h. The acid precipitate was collected by centrifugation at 15,000 × g for 30 min and re-suspended in distilled water and adjusted to pH 7.0 to make most of the precipitate dissolved and centrifuged again. For the extraction of biosurfactant compounds, 50 ml of chloroform- methanol (2:1, v/v) was added to 500 mg of the dry product and incubated in a rotatory shaker at 250 rpm, 30ºC for 15 minutes. The mixture was filtrated using a 0.45 µm Millipore membrane (Thaniyavarn et al. 2006). The filtrate was lyophilized, weighed for quantification and then used for antimicrobial activity tests.
Antimicrobial assay
The antimicrobial activity of the produced biosurfactant was studied against different Gram positive, Gram negative bacteria and yeast (Table 1). The identified strains were obtained from the Microbial Culture Collection (MIRCIN) at Faculty of Agriculture, Ain Shams University. The antimicrobial activity was evaluated by the agar disc diffusion method (Bauer et al. 1966). Sterile discs (0.6 cm) soaked with the biosurfactant solution in methanol was assayed on the surface of an nutrient agar and malt extract medium for bacteria and yeast, respectively inoculated with the tested microorganism. After incubation period for 24 h at 37± 2 °C and for 48 h at 25 ± 2 °C for bacteria and yeast, respectively, the diameter of inhibition zones was measured (Bradshaw 1992). Negative controls were prepared using the same solvents as employed to obtain the extract. As positive controls, ofloxacin (5 μg, Oxoid) was used for Gram-positive bacteria, cefaperazonesulbactam (10 μg, Oxoid) for Gram-negative bacteria and amphotericin B (30 μg, Sigma) for Candida albicans. To ensure that the results were reproducible, the average of three independent measurements was taken.
MIC determination
The minimum inhibitory concentration (MIC) is defined as the lowest concentration of biosurfactant at which no visible growth could be observed after incubation for the required time (Ericsson and Sherris 1971). MIC for S. aureus ATCC 25928 was determined by the broth dilution method as described Evans et al. (1996).
Effect of biosurfactant on growth and intracellular components of Staphylococcus aureus ATCC 25928
Erlenmeyer flasks (500 ml volume) containing 100 ml of the nutrient broth medium were inoculated with a suspension of S. aureus cells (O.D600 = 107 cells/ ml). Different concentrations of biosurfactant (0, 6, 12, 24, 36 and 48 µg / ml) were added. The inoculated flasks were incubated on a rotary shaker (150 rpm) at 37 oC±2°C for 12 h. Ten milliliter samples were withdrawn at 2 h intervals (after 0, 2, 4, 6, 8, 10 and 12 h) to elucidate the effect of biosurfactant on the bacterial growth and intracellular components.
Cell growth measurement
Bacterial growth was determined by measuring the absorbance at 600 nm (OD600) by a spectrophotometer (UV-VIS Double Beam PC, Labomed INC) (Kim et al.1999).
Fractionation and quantitative estimation of intracellular components
1) Acid - soluble phosphorous compounds
Ten milliliter samples of biosurfactant treated and untreated cultures (control) were withdrawn at 2 h intervals (after 2, 4, 6, 8, 10 and 12 h). The cells were collected by centrifugation, washed twice with ice-cold saline solution (0.9 % NaCl) and extracted twice with 2.5 ml of 5% ice-cold trichloroacetic acid (TCA). The suspension was finally centrifuged at 6000 rpm for 10 minutes and the combined TCA extracts were used for the determination of acid-soluble phosphorous according to the method described by Toribarn et al. (1956).
2) Total lipids
The residue after the removal of the acid-soluble phosphorous was extracted three times with 5.0 ml mixture of chloroform: methanol (2:1, v/v) as described by Bligh and Dyer (1959).The combined extracts were used for the determination of total lipids according to Knight et al. (1972).
3) Total proteins
The dilapidated cells were incubated with 2.0 ml 1N KOH at 37 °C for 20 h. One milliliter of the product was saved for protein determination as described by Lowry et al. (1951) using bovine serum albumin as standard.
4) RNA
The remaining portion of the samples (1.0 ml) after hydrolysis by 1 N KOH was subjected to extraction of RNA and DNA fractions. For this, 0.4 ml of 6N HCl was added to each sample, then the solution was completed with the same volume of 10 % TCA. After centrifugation, the supernatant was used for RNA determination as described by Merchant and Kahn (1969).
5) DNA
The residue after the extraction of RNA was hydrolyzed with 5.0 ml of 5% TCA at 90 °C for 30 minutes, cooled and centrifuged at 6000 rpm. The residue was washed once with 2.0 ml of 5% TCA and the supernatants were combined to form DNA fraction, then the DNA content was measured as described by Dische and Chargaff (1955).
Statistical analysis
Results are presented as mean value ± standard deviation (SD). The Microsoft Excel 2003 and SAS 9.1.3 statistical program were used for data analysis.
RESULTS AND DISCUSSION
Ten bacterial strains were isolated from the oil-contaminated soil samples with different degrees of oil contamination collected from different plots around the oil wells of western desert of Egypt. Depending on qualitative drop-collapse method of Youssef et al. (2004), the most efficient biosurfactant- producing bacterial strain, identified as B. licheniformis strain M104, was selected for further studies (data not shown).
The production of biosurfactant depends on the type of carbon source present in the medium (Davis et al.1999; Adamczak and Bednarsk 2000). In this study, whey was used for biosurfactant production. Whey from the dairy industries has been reported previously as a cheap source for good microbial growth and biosurfactant production (Patel and Desai 1997; Dubey and Juwarkar 2001; 2004). These studies showed that whey wastes might be comparatively better substrates for biosurfactant production at the commercial scale than the synthetic medium. Furthermore, the potential use of dairy wastewaters provides a stratagem for the management of efficient dairy wastewater.
One useful property of many biosurfactants that has not been reviewed extensively is their antimicrobial activity (antibacterial, antifungal and antiviral). Other medically relevant uses of the biosurfactants include their role as anti-adhesive agents to pathogens, making them useful for treating many diseases and as therapeutic agents (Singh and Cameotra 2004). In the present study, the lipopeptide biosurfactant produced by B. licheniformis strain M104 exhibited interesting antimicrobial activities. Table 1 showed that all the tested microorganisms were sensitive to the biosurfactant, except a Gram-positive bacterium (L. monocytogenes) and a Gram-negative bacterium (K. pneumoniae), which showed resistant to biosurfactants. Several lipopeptide biosurfactants produced by B. licheniformis have been shown to have antimicrobial activity (Jenny et al. 1991; Fiechter 1992; Yakimov et al. 1995).
S. aureus was the most sensitive strain to biosurfactant. These results were in accordance with Singh and Cameotra (2004) who reported that lipopeptide produced by B. subtilis was active against several microorganisms, especially S. aureus. The management of S. aureus infections is a major problem due to wide spread resistance to beta-lactams and glycopeptides (Heinemann 1999).
The produced biosurfactant showed an antifungal activity against C. albicans. Thimon et al. (1995) reported that the biosurfactant produced by B. subtilis had an antifungal effect on yeast cells. Nevertheless, the lipopeptide surfactants produced by Bacillus genus present a great potential for biotechnological and biopharmaceutical applications due their biological properties (Singh and Cameotra 2004).
Sheppard et al. (1991) showed that various interesting biological properties of lipopeptide biosurfactants were presumed to be the result of interactions with the membranes of target cells. One explanation of the antimicrobial effect of the biosurfactants is the adhering property of the biosurfactants to the cell surfaces caused deterioration in the integrity of cell membrane and also breakdown in the nutrition cyle (Hingley et al. 1986). Another explanation is the amphiphilic structures of biosurfactants, insertion of fatty acids components of biosurfactants into a cell membrane caused an increase in the size of the membrane and significant ultrastructural changes in the cells such as ability of the cell to interiorize plasma membrane. Alternatively, it is possible that the insertion of the shorter acyl tails into the cell membrane causes a disruption between the cytoskeletal elements and the plasma membrane, allowing the membrane to lift away from the cytoplasmic contents (Desai and Banat 1997).
However, the ways in which the biosurfactants affect the membrane integrity differ. For example, Thimon et al. (1995) suggested that the lipopeptide biosurfactant was thought to disrupt the plasma membranes of the cells by the accumulation of intra membranous particles in the cells and increasing the electrical conductance of the membrane. On the other hand, Carrillo et al. (2003) reported that the lipopeptide biosurfactant increased the membrane permeability though the interaction with the cell membrane phospholipids.
The minimum inhibitory concentration (MIC) of the biosurfactant for S. aureus ATCC 25928 was 6 μg / ml. It was worthy to note that the produced lipopeptide biosurfactant showed a good antimicrobial activity in comparison to the rhamnolipid produced by P. aeruginosa (32 µg / ml, Abalos et al., 2001) and (8 µg/ml, Benincasa et al.2004). Five concentrations of the biosurfactant (6, 12, 24, 36 and 48 µg / ml) were selected, as multiplication of MIC, for studying their influences on the cell growth and intracellular components of the most sensitive bacteria, S. aureus ATCC 25928.The antimicrobial effect of lipopeptide biosurfactant was time and concentration dependent. It showed the maximum inhibitory effect on the cell growth and intracellular components at 48 µg / ml after 12 h in comparison with the control cells. Jones et al. (1996) reported that the antimicrobial agents that affect the growth and multiplication of certain types of cells may interact with various targets (chemo-receptors) in the sensitive cell. Theoretically, these targets might be numerous but with decreasing the concentrations of the antimicrobial agent; the number of targets also decreased.
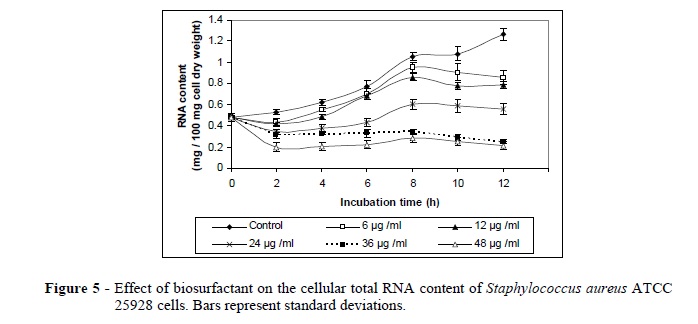
The results of the effect of the produced biosurfactant on the S. aureus ATCC 25928 cells, including the growth rate, acid soluble phosphorous, total lipids, total proteins, RNA and DNA are presented in Figures 1-6.The maximum reduction of acid soluble phosphorous (53.06 %), total lipid (90.47 %), total proteins (53.43%), RNA (83.29 %) and DNA (48.50%) were reached after 12 h using 48 µg/ml of the biosurfactant. The biosurfactant exerted a significant decrease in the total lipid content of S. aureus ATCC 25928 cells after 12 h as compared to the normal control cells (Fig 3). This decrease could be due to the effect exerted by the biosurfactant on the permeability of cell membrane or on the level of double layer of bacterial membrane (Novo et al. 2000).
Vander (1985) reported that the alternation in the fatty acid contents could be due to the disturbance in membrane permeability resulting from the direct interaction of the biosurfactant with the lipids, which caused inhibition of the membrane bounded enzyme and leakage of intracellular components. In the same regard, Cameotra and Makkar (2004) reported that the antimicrobial biosurfactant could disturb the membrane structure through the interaction with phospholipids as well as membrane proteins.
A decrease in total protein content of S. aureus ATCC 25928 cells after the treatment with the biosurfactant could be due to the inhibition of protein synthesis (Fig 4). It might bind to the 30S ribosome subunit and thus prevented the association of aminoacyl-tRNA with the bacterial ribosome as reported by Schnappinger and Hillen (1996). Singh et al. (2002) suggested that the antimicrobial biosurfactant prevented the protein synthesis by inhibition of the peptidyltransferase in binding mainly the 23S rRNA in the 50S subunit of the bacterial ribosome. The decrease in the lipids content of S. aureus cells after treating with the biosurfactant had a relation with the disturbance in the proteins content. This could be attributed to the property of expanding membrane protein causing the conformational changes of lipid and protein content (Price et al. 2001).
The biosurfactant also exhibited a decrease in the total RNA and DNA contents of S. aureus cells. This could be due to the loss of intracellular components through the damaged cell wall (Figs. 5, 6). Volk et al. (1996) reported that the bactericidal agent formed the complexes with guanine residues in helical DNA. It also prevented the RNA polymerase on the DNA template. At higher concentrations, DNA replication also was inhibited. It could also be caused by selectively inhibiting an enzyme (DNA gyrase) needed for the replication of DNA (Drlica and Hooper 2003). The bactericidal agent binds to the ß subunit of the DNA-dependent RNA polymerase and inhibits the initiation of transcription by preventing the synthesis of RNAs larger than dinucleotides (Chaisson 2003).
In conclusion, results of the present study demonstrated that the biosurfactant produced by B. licheniformis strain M104 presented a great potential for the biotechnological and biopharmaceutical applications due to their biological properties.
Received: November 22, 2011;
Revised: April 16, 2012;
Accepted: December 17, 2012.
- Abalos A, Pinazo M, Infante R, Casals M, Garcia F , Manresa A. Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes. Langmuir. 2001;17: 1367-1371.
- Adamczak M, Bednarsk W. Influence of medium composition and aeration on the synthesis of biosurfactants produced by Candida antractica Biotechnol Lett. 2000; 22: 313-316.
- Banat IM. Characterization of biosurfactants and their use in pollution removal-state of the art. Acta Biotechnol.1995; 15: 251-267.
- Bauer AW, Kirby WM, Sherris JC, Ture KM. Antibiotic susceptibility testing a standardized single disk method. Am J Clin Path. 1966 ; 45: 493-496.
- Benincasa M, Abalos A, Oliveira I , Manresa A. Chemical structure, surface properties and biological activities of the biosurfactant produced by Pseudomonas aeruginosa LBI from soapstock. Ant Van Lee. 2004; 85:1-8.
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959; 37: 911-917.
- Bodour AA, Drees KP, Maier RM. Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid southwestern soils. Appl Environ Microbiol. 2003; 69: 32803287.
- Bradshaw LJ. Laboratory Microbiology. Fourth Edition. USA; 1992. 13-55.
- Cameotra SS, Makkar RS. Recent applications of biosurfactants as biological and immunological molecules. Curr Opin Microbiol. 2004 ; 7: 262-266.
- Cao XH, Cai P, Li F, Wang CL , Lu MF. Medium optimization for lipopeptide produced by Bacillus natto TK-1 using response surface methodology. China Biotechnol . 2007; 27: 59-65.
- Carrillo C, Teruel JA, Aranda FJ , Ortiz A. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim Biophys Acta. 2003; 611: 91-97.
- Chaisson RE. Treatment of chronic infections with rifamycins: is resistance likely to follow? Antimicrob Agents Chemother. 2003; 47: 3037-3039.
- Davis DA, Lynch HC, Varely J. The production of surfactin in batch culture by Bacillus subtilis ATCC 21332 is strongly influenced by the condition of nitrogen metabolism. Enzyme Microb Technol. 1999; 25: 322-329.
- Desai JD, Banat IM. Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev. 1997; 61: 47-56.
- Deziel E, Paquette G, Villemur R, Lepine F, Bisaillon JG. Biosurfactant production by a soil Pseudomonas strain growing on polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 1996; 62:19081912.
- Dische Z. Chargaff E. The Nucleic Acids. In: Davidson IN, editor. Academic press, New York; 1955. p. 130-157.
- Drlica K , Hooper DC . Mechanisms of quinolone action. In: Hooper DC, Rubinstein E, editor. Quinolone antimicrobial agents, 3rd edn. Am Soc Microbiol; 2003. p. 19-40.
- Dubey K, Juwarkar A. Distillery and curd whey wastes as viable alternative sources for biosurfactant production. World J Microbiol Biotechnol. 2001; 17: 6169.
- Dubey K, Juwarkar A. Determination of genetic basis for biosurfactant production in distillery and curd whey wastes utilizing Pseudomonas aeruginosa strain BS2. Indian J Biotechnol. 2004; 3: 7481.
- Ericsson H , Sherris JC. Antibiotic sensitivity testing. Report of an international collaborative study. Act Pathol Microbiol Sc and B Suppl. 1971; 217-221.
- Evans WC. Pharmacopoeial and related drugs of biological origin. In: Trease and evan pharmacognosy. London: Saunders WB Co. Ltd; 1996.
- Fiechter A. Biosurfactants: moving towards industrial application. Trends Biotechnol.1992; 10: 208-217.
- Heinemann JA. How antibiotics cause antibiotic resistance. Drug Discov Today. 1999; 4: 72-79.
- Hingley ST, Hastie AT, Kueppers F. Effect of ciliostatic factors from Pseudomonas aeruginosa on rabbit respiratory cilia. Infect Immun. 1986; 51: 254-258.
- Jenny K, Kappeli O, Fiechter A. Biosurfactants from Bacillus licheniformis, structural analysis and characterization. Appl Microbiol Biotechnol.1991; 36: 5-13.
- Jones M, Comer I, Cunha B. Tetracyclines. In: Raven H, editor. J. Antimicrobial Therapy, New York; 1996. p. 219-234.
- Joshi S, Bharucha C, Jha S, Yadav S, Nerurkar A, Desai A. Biosurfactant production using molasses and whey under thermophilic conditions. Bioresource Technol. 2008; 99: 95199.
- Khire JM , Khan MI . Microbes and the sub-surface environment. Enzyme Microbiol Technol. 1994; 16: 258-259.
- Kim MS, Yoon BD, Choung DH , Katsuragi OH , Tani Y. Characterization of a biosurfactant manosylerythritol lipid produced from Candida sp. SY.16. Appl Microbiol Biotenol. 1999; 52:713-721.
- Knight JA, Anderson S, Rawle JM. Sulfo-phosphovanillin reaction for the estimation of total serum lipids. J Clin Chem, 1972; 18: 199-209.
- Lowry OH, Rosebrough N, Farr AL , Randall R.J. 1951, Protein measurement with Folin phenol reagent. J Biol Chem. 1972; 193: 265-275.
- Makkar RS, Cameotra SC. Biosurfactant production by microorganisms on unconventional carbon sources. J Surf Deterg. 1999; 2: 237241.
- Merchant DJ, Kahn RH. Hand book of cell organ culture. In: Murphy WH, editor. Second edition, Bugass Mincecopolis; 1969.
- Mukherjee S, Das P, Sen R. Towards commercial production of microbial surfactants. Trends in Biotechnol. 2006; 24: 509-515.
- Novo D, Perlmatter N, Hunt R, Shapiro H. Multiparameter flow cytometric analysis effect on membrane potential, membrane permeability and bacterial counts for Staphylococcus aureus and Mycrococcus luteus Antimicrob Agent Chemother. 2000; 44: 827-838.
- Patel RM, Desai, AJ. Biosurfactant production by Pseudomonas aeruginosa GS3 from molasses. Lett Appl Microbiol. 1997; 25: 9194.
- Peypoux F, Bonmatin JM, Wallach J . Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol. 1999; 51: 553-563.
- Price AC, Choi KH, Heath RJ, Li Z, White SW , Rock CO. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism. J Biol Chem. 2001; 276: 6551-6559.
- Ramadan EM, Kheiralla ZM, Fouad MA, El Tayeb T S, Gomaa EZ. Optimization of biosurfactant production by Bacillus licheniformis isolated from oil contaminated Egyptian soil. Egypt J Microbiol. 2011; 45, In press.
- Rocha C, San-Blas F, San-Blas G , Vierma L. Biosurfactant production by two isolates of Pseudomonasaeruginosa World J Microbiol Biotech. 1992; 8: 125-128.
- Schnappinger D , Hillen W. Tetracyclines, antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996; 165: 359-369.
- Sepahy AA, Assadi MM, Saggadian V, Noohi A. Production of biosurfactant from Iranian oil fields by isolated Bacilli. Int J Environ Sci Technol. 2005; 1: 287-293.
- Sheppard JD, Jumarie C, Cooper DG , Laprade R. Ionic channels induced by surfactin in planar lipid bilayer membranes. Biochem Biophys Acta. 1991; 1064: 13-23.
- Singh A, Van Hamme JD , Ward OP. Surfactants in microbiology and biotechnology, Part 2. Application aspects Review Article. Biotechnol Adv. 2007; 25: 99-121.
- Singh KV, Weinstock GM, Murray BE. An Enterococcus faecalis ABC homologue Lsa, is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 2002; 46: 1845-1850.
- Singh P, Cameotra S. Potential applications of microbial surfactants in biomedical sciences. Trends Biotechnol. 2004; 22: 142146.
- Thaniyavarn J, Chongchin A, Wanitsuksombut N, Thaniyavarn S, Pinphanichakarn P, Epipatpiboon N, Morikawa M , Kanaya S. Biosurfactant production by Pseudomonas aeruginosa A41 using palm oil as carbon source. J Gen Appl Microbiol. 2006; 52: 215222.
- Thimon L, Peypoux F, Wallach J , Michel G. Effect of the lipopeptide antibiotic iturin A, on morphology and membrane ultrastructure of yeast cells. FEMS Microbiol Lett.1995; 128: 101-106.
- Toribarn JR, Chen PS , Warner H. Assay of inorganic phosphate, total phosphate, and phosphatases. In: Elizabeth FN, Vector G, editor. Method Enzymol, Vol III; 1956. p. 115-116.
- Vander BH. Biochemical targets for antifungal azole derivatives, hypothesis the mode of action. Curr Top Med Mycol. 1985; 1: 313-351.
- Van Hamme JD, Singh A , Ward OP. Physiological aspects: Part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol Adv. 2006; 24: 604-620.
- Volk W, Gibhardt B, Hammarskjolg M, Kadner R. Essential of Medical Microbiology. In: Lippencott R, editor; 1996. p. 153-256.
- Yakimov MM, Timmis KN, Wray V, Fredrickson H L. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl Environ Microbiol. 1995; 61: 17061713.
- Youssef OH, Duncan KE, Nagle DP, Sava KN, Knapp RM , McInerney MJ . Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods. 2004; 56: 339 347.
Publication Dates
-
Publication in this collection
24 May 2013 -
Date of issue
Apr 2013
History
-
Received
22 Nov 2011 -
Accepted
17 Dec 2012 -
Reviewed
16 Apr 2012