ABSTRACT
Mungbean (Vigna radiata (L.) Wilczek) also known as green gram is an important source of protein in the category of food legumes. In the present study, SSR marker is used to analyze the genetic diversity amongst 23 genotypes of mungbean. Out of a total of 10 primers used for SSR analysis revealed generation of 15 alleles. The number of alleles per locus ranged from one (CEDG006, CEDG010, CEDG050, CEDG088, CEDG092 and CEDG232) to three (CEDG 214), with an average of 1.5 allele per primer. The index for expected heterozygosity was 0.29 ranging from 0.15 to 0.49 revealed a deficit in heterozygosity. The size of amplification products varied in case of each primer and the range was found to be 100 bp to 190 bp. 13 out of 15 alleles were found polymorphic. The average PIC value of SSR marker was found to be 0.205. The value of Jaccard’s similarity coefficient had ranged from 0.28-1.00 with an average value of 0.64. The dendrogram constructed on SSR molecular marker data through UPGMA method and PCA using average linkage, had enabled grouping of the genotypes into three main clusters. Clustering pattern based on SSR marker data clearly indicated the narrow genetic base of mungbean genotypes that emphasizes the need to explore and exploit more number of germplasm from additional source to study genetic variation in mungbean for genetic improvement. The results indicated the marked usefulness of SSR in the assessment of genetic diversity in mungbean crop.
Key words:
Genetic diversity; Mungbean; SSR Marker assisted selection
INTRODUCTION
Mungbean is also referred to as green gram, golden gram, green bean, mash bean, green soy and its botanical name is Vigna radiata L. (Wilczek). It belongs to the family Fabaceae (Leguminoceae). It is an important legume crop grown in 2.75 million hectare with a production of 1.19 million tones11 Economic Survey. Ministry of Finance, Department of Economic Affairs.Economics Division Government of India, New Delhi. 2012-2013.. It is a diploid with the 2n= 22 chromosome number22 Arumuganathan K and Earle ED . Nuclear DNA content of some important plant species. Plant Molecular Biology Report .1991; 9: 208-218.. Mungbean not only provides proteins but also maintains soil fertility by fixing atmospheric nitrogen (30-50 kg/ha). It is grown primarily as an intercrop with wheat, maize, potato, etc during kharif and as monocultures during rabi and zaid. It is grown primarily as an intercrop with wheat, maize, potato, etc during kharif and as monocultures during rabi and zaid.
On account of its short duration, photo-insensitivity and dense crop canopy, it assumes special significance in crop intensification, diversification, and conservation of natural resources as well as sustainability of the production system. Mungbean is an excellent source of high quality protein, Vitamin C, Vitamin B, folate and mineral nutrients eg. calcium, magnesium, phosphorous, potassium etc33 Saini M, Singh S, Hussain Z and Yadav A. RAPD analysis in mungbean (V. radiata (L.) Wilczek): I. Assessment of genetic diversity. Indian Journal of Biotechnolog. 2010;9: 137-146.. The low productivity of this crop can be attributed to narrow genetic base resulting in low yield potential and susceptibility to biotic and abiotic stresses44 Poehlman JM . The Mungbean. Oxford and IBH Publication, New Delhi. 1991.. Although germplasm collection within India is quite large, much diversity has not been reported on the basis of morphological characters. Therefore, an urgent need to identify genetic divergence based on molecular analysis is an essential pre-requisite for further utilization in crop improvement programmes.
SSR markers have been widely used for plant genetic analysis because of their abundance, co-dominant inheritance, high polymorphism, reproducibility, and ease of assay by PCR 55 Kuleung C, Baenziger PS and Dweikar I. Transferability of SSR markers among wheat, rye, and triticale. Theoretical and Applied Genetics.2004; 108(6): 1147-1150.,66 Xie RJ, Zhou J, Wang GY, Zhang SM, Chen L and Gao ZS. Cultivar identification and genetic diversity of Chinese bayberry (Myrica rubra) accessions based on fluorescent SSR markers. Plant Molecular Biology Reporter. 2011; 29(3): 554-562.. SSR are rapidly becoming a preferred type of DNA marker over RAPD and ISSR in its use for germplasm analysis because SSR are locus specific, widely dispersed throughout the genome, highly polymorphic due to variation in the repeat units and highly informative because of co-dominant nature and are a powerful tool for genetic analysis. SSRs are thus markers of choice for genetic studies because of their hyper variability, co-dominant nature, locus specificity and high reproducibility. Chen et al., 2015 developed 500 novel expression sequences tag (EST) based SSRs (eSSRs) and genomic SSRs (gSSRs) from mung bean transcriptome and genome sequences for diversity evaluation77 Chen H, Qiao L, Wang L, Wanga L, Blair MW and Cheng Z. Assessment of genetic diversity and population structure of mung bean (Vigna radiata) germplasm using EST-based and genomic SSR markers. Gene . 2015; 566 (2): 175-183. In recent years, mungbean gSSRs and eSSRs detection has accelerated through NGS/deep sequencing however validation of these SSRs are yet to be studied88 Chen H, Wang L, Wang S, Liu C, Blair MW and Cheng Z. Transcriptome Sequencing of Mung Bean (Vigna radiate L.) Genes and the Identification of EST-SSR Markers. PLoS ONE. 2015; 10(4): e0120273.. In the present the genetic diversity of 23 genotypes of mungbean were estimated using SSR molecular markers.
MATERIAL AND METHODS
PLANT MATERIALS AND DNA EXTRACTION
In the present investigation seeds of twenty three genotypes of green gram were procured from Agriculture Research Station (ARS), Durgapura, Jobner Agriculture University, Jaipur. Source details of the materials used are given in Table 1. Genomic DNA was isolated and purified from young leaves of 21-28 days99 Doyle JJ and Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochemical Bulletin.1987; 19: 11-15. and stored at -200C for further analysis.
SSR PRIMERS AMPLIFICATION
Forty SSR primers were screened in the mungbean germplasms, out of which 10 primers generated polymorphic and reproducible banding pattern and were selected for final analysis. PCR amplification was carried out in a 20 μL reaction volume containing 200 µM of dNTP mix, 1.5 mM MgCl2, 1U of Taq polymerase, 1X of reaction buffer, 0.5 µM forward and reverse primer and 25 ng genomic DNA. The amplification was performed in an Eppendrof Master cycler, with reaction conditions programmed as initial pre-denaturation at 94 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 57.3°C to 60.85°C for 1 min, and extension at 72 °C for 45 sec. A final extension was done for 10 min at 72 °C with a hold temperature of 4°C. The above obtained PCR products were mixed with 2 μl of 6X loading dye (Bromophenol blue) and were loaded on 4 per cent metaphor agarose gel prepared in 1X TAE buffer containing ethidium bromide (10 mg/ml) concentration of 3µl/100ml1010 Sambrook J, Fritschi EF and Maniatis T . Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York. 1989.. Electrophoresis was carried out and gel was visualized on a UV-transilluminator and photographed using gel documentation system.
DATA ANALYSIS
Amplified bands generated by SSR -PCR amplification were scored based on the presence (1) or absence (0) of bands for each primer and were used to calculate a genetic similarity matrix employing the Jaccard’s similarity coefficient using NTSYS-pc version 2.11111 Rohlf FJ. NTSYS-PC. Numerical taxonomy and multivariate analysis system.Exeter Software, New York. 1989.. Cluster analysis was performed on molecular data using the unweighted pair group method using arithmetic means (UPGMA) algorithm1212 Sneath PHA and Sokal RR. Numerical Taxonomy. San Francisco, Freeman. 1973., from which dendrograms depicting the similarity among varieties were drawn and plotted [PIC] using NTSYS-pc1111 Rohlf FJ. NTSYS-PC. Numerical taxonomy and multivariate analysis system.Exeter Software, New York. 1989..To measure the polymorphism information content (PIC) of SSR marker system the following formula was applied : PIC=1-nΣpi22 Arumuganathan K and Earle ED . Nuclear DNA content of some important plant species. Plant Molecular Biology Report .1991; 9: 208-218. where, n= total number of allele detected for a locus of a marker; pi= frequency of the 1st allele1313 Smith JSC, Chin EL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S and Zeigle J . An evaluation of the utility of SSR loci as molecular markers in maize (Z. mays L.):comparisons with data from RFLPs and pedigree. Theoretical and Applied Genetics.1997; 95: 163-173.. The effective number of alleles, Nei’s genetic diversity/ expected number of hetrozygosity and Shannon's Information index were computed using POPGENE software 1414 Yeh FC, Boyle TJB. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian J. Bot.1997; 129:157-163.. A two dimensional and three dimensional principal component analysis (PCA) was constructed to provide another means of testing the relationship among genotypes using the EIGEN programme NTSYS-pc version 2.02 1111 Rohlf FJ. NTSYS-PC. Numerical taxonomy and multivariate analysis system.Exeter Software, New York. 1989..
RESULTS AND DISCUSSION
Genetic diversity among 23 mungbean genotypes: Ten out of the forty SSR primers used in the present investigation showed amplification in all the genotypes. Out of ten SSR primers, eight showed good polymorphism. A total of 15 alleles were detected in the 23 mungbean genotypes using 10 SSR markers, out of which 13 were polymorphic with an average polymorphism of 86.66 per cent. The total number of alleles observed for each primer was recorded separately and percentage polymorphism calculated subsequently (Table 2). Overall size of PCR amplified products ranged between 100 bp to 190 bp. PIC values, a measure of the allelic diversity, ranged from 0.00 to 0.45 with an average of 0.205. The total numbers of alleles observed for each primer were recorded separately and are shown in Table 2 .The DNA banding pattern of individual plant DNA samples from the 23 genotypes of mungbean. After amplification with SSR primers are depicted in Fig.1.The similarity indices between genotypes ranged from 0.28 to 1.00 or the genetic diversity ranged from 0 to 72%.
SSR profiles ofVigna radiata L. genotypes generated through (a) CEDG050 (b)CEDG088. G1-G23 represents G1-PUSA672; G2-AKM962; G3-UPM02-18; G4-ML-729; G5-EC-398885; G6-IPM-02-1; G7- IPM-02-03; G8-IPM02-14; G9-IPOI-1539; G10-RMG-62; G11-PDM-288; G12-RMG-353; G13-PRETEEKHSA-NEPAL; G14-MEHA; G15-PANT; G16-ASHA;G17-MG331;G18-GM9925, G19-IC-393407; G20-DRA-24; G21-SAMRAT; G22-HUM-1 and G23-HUM-12. M1-100 bp DNA Ladder and M2- 1000 bp DNA Ladder
The average similarity across all 23 genotypes was found out to be 0.64, showing that genotypes were moderately similar to each other. Maximum similarity value of 1.00 was observed between genotypes IPOI-1539 and ML-729; PANT and ML-729; IPOI-1539 and IPM 02-01; PANT and IPOI-1539; IC-393407 and IPM 02-14; DRA-24 and IPMO2-14;HUM-12 and IPMO2-14;PANT and IPOI-1539; DRA-24 and IC-393407; HUM-12 and IC-393407; HUM-12 and DRA-24, respectively. Genotypes PDM-288 and EC-398885, as well as MEHA and EC-398885 were found to be genetically diverse with minimum similarity value of 0.40.
Similar polymorphism results have been shown previously by Singh, et al. (2012) where authors estimated genetic diversity of the induced variation among 30 mutant lines and parent genotype (Pusa 9072)1515 Singh SK, Lavanya GR, Bhat KV, Babu GS, Arya L, Verma M, Hussain Z, Roy S, Rathi RS and Mishra AK. Microsatellite Markers Revealed Genetic Diversity in Mungbean Mutant Lines. Indian Journal of Hill Farming. 2012; 25(1): 38-43.. The size of amplification products varied in case of each primer and the range was 50 bp to 250 bp. A total of 98% polymorphism was observed through STMS markers among the mutants. Gupta et al. (2013) had studied 30 SSR markers in mungbean genotypes, out of which 19 (63%) markers exhibited polymorphism1616 Gupta S, Gupta DS, Anjum KT, Pratap A and Kumar J. Transferability of simple sequence repeat markers in black gram (V. mungo L. Hepper). Australian Journal of Crop Science.2013; 7(3): 345-353.. Sherawat et al. (2014) studied thirty eight novel microsatellite markers (SSRs) specific to candidate genes for detection of genetic variations in 12 mungbean genotypes1717 Sehrawat N, Bhat KV, Kaga A, Tomooka N, Yadav M and Jaiwal PK. Development of new gene-specific markers associated with salt tolerance for mungbean (Vigna radiata L.Wilczek). Spanish Journal of Agricultural Research. 2014;12 (3): 732-741. and reported the maximum similarity value of 0.56 between two salt-tolerant cultivated genotype (PLM562 and PLM380) of mungbean.
GENETIC VARIATION OF SSR LOCI WITHIN MUNGBEAN GENOTYPES
Table 2 represents individual locus statistics for SSR analysis: A total of 15 alleles were observed at the 10 SSR loci. Allelic diversity at SSR loci varied greatly from one locus to another, and was high on average. The number of alleles “A” ranged from 1 to 3 with an average of 1.5 “Ne” was 1.47 on average and ranged from 1.40 to 1.88. “He” exhibited a range of variation from 0.15 to 0.49 with a mean of 0.29. “I” averaged 0.46 and ranged from 0.29 to 0.69. The most polymorphic loci were identified in CEDG50 followed by CEDG10, CEDDG6, CEDG88, and CEDG92 that revealed a high level of genetic variation.
The similar results for the expected heterozygosity (He) also referred as Nei’s gene diversity for the polymorphic SSR markers were reported by Gupta et al., (2013) which was ranged from 0.03 to 0.63, indicated the presence of low genetic diversity in the Indian mungbean1818 Gupta SK, Bansal R, Vaidya UJ and Gopalakrishna T. Assessment of genetic diversity at molecular level in mungbean (Vigna radiata (L.) Wilczek). Journal of Food Legumes. 2013; 26(3 & 4): 19-24.. The presence of very low observed hetrozygosity in Indian mungbean genotypes is expected since it is a self pollinated crop.
ASSOCIATIONS AMONG INDIVIDUALS OF MUNGBEAN GENOTYPES
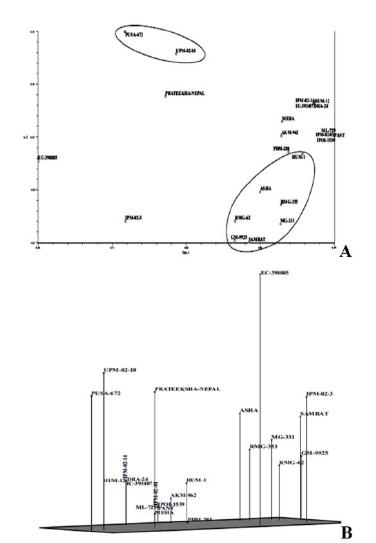
The UPGMA analysis based cluster tree analysis divided the 23 mungbean genotypes into three major clusters at a similarity coefficient of 0.80 (Fig. 2). Cluster I included two genotypes PUSA-672 and UPM-02-18 which were found similar to each other at a similarity coefficient of 0.79. Cluster II included eleven genotypes, viz., AKM-962, ML-729, IPM-02-01, IPOI-1539, PANT, IPM-02-14, IC-393407, HUM-12, DRA-24, MEHA and PDM-288 again found similar to each other at a similarity coefficient of 0.79. Cluster III consisted of seven genotypes viz., RMG-62, RMG-353 SAMRAT, HUM-1, ASHA, MG-331, and GM-9925 that were found similar to each other at a similarity coefficient of 0.88. Three genotypes viz.. PRATEEKSHA-NEPAL, IPM-02-3 and EC-398885 were out grouped from all the clusters thus indicating that they are divergent. Two and three dimension principal component analysis based on SSR data (Fig. 3) showed similar clustering pattern of the 23 genotypes as evident from cluster tree analysis. Similarity coefficients ranged from 0.50 to 0.97, indicative of an average degree of variation. Similar to the UPGMA cluster tree, most of the genotypes were found to be grouped into three main groups.
Dendrogram constructed with UPGMA clustering method among 23 genotypes of V. radiata L. using SSR markers.
(a) Two Dimensional Principal Component Analysis (b) Three Dimensional Principal Component Analysis based on Euclidean Cluster Analysis Using SSR Markers.
Similar clustering were also observed by Gupta et al., (2013)18 through UPGMA analysis based on 19 polymorphic SSR markers grouped the 29 mungbean genotypes into three main clusters with Dice’s analysis, the similarity coefficient ranging from 0.31 to 1.0. Dikshit et al. found that the SSR marker was more efficient in detecting genetic variability among all Vigna species1919 Dikshit HK, Jhang T, Singh NK, Koundal KR, Bansal KC, Chandra N, Tickoo JL and Sharma TR . Genetic differentiation of Vigna species by RAPD, URP and SSR markers. Biologia Plantarum. 2007; 51(3): 451-457.. The results of the PCA analysis were also congruent well with Gupta et al. (2013).
The traditional method of SSR marker development usually includes the construction of genomic library, cloning and sequencing, which is costly and labor intensive. With the advancement in sequencing technologies viz. RNA sequencing with NGS technology has provided an effective way to develop EST-SSRs, which makes them a valuable resource of these SSRs for future genetic studies. Liu et al. (2016) identified 3788 expressed sequence tag-simple sequence repeat (EST-SSR) motifs in mungbean which could be used as potential molecular markers upon validation2020 Liu C, Fan B, Cao Z, Su Q, Wang Y, Zhang Z, Wu J and Tian J. A deep sequencing analysis of transcriptomes and the development of EST-SSR markers in mungbean (Vigna radiata). Journal of Genetics. 2016; 95: 527-535.. Validation of molecular markers e.g. SSRs are one step toward providing precise genetic information for future breeding program and accurately characterize the genotype portraying for the extent of genetic diversity2121 Goyal P, Jain R, Kachhwaha S and Kothari SL. Assessment of genetic diversity in Pithecellobium dulce (Roxb.) Benth. germplasm using RAPD and ISSR markers. Trees. 2015; 29: 637-653. The present study discusses the validation of such SSRs and polymorphic SSRs can further be employed for crop improvement.
CONCLUSION
Mungbean is an important leguminous crop plant widely cultivated throughout the southern half of Asia. In the present study, SSR markers detected a moderate level of genetic diversity in mungbean. Each polymorphic SSR marker detected one to three alleles with an average of 1.5 alleles per locus. The present study revealed that SSR markers are very effective in detecting the genetic variation in mungbean and could be used for genetic analysis in mungbean. The result showed the presence of narrow genetic diversity in Indian mungbean genotypes and would help the mungbean breeders in selection of suitable parents for breeding purposes and genetic mapping studies.
ACKNOWLEDGEMENT
Gunnjeet thanks UGC, New Delhi, for awarding SRF under the Maulana Azad National Fellowship for Minority Students. Dr. N. S. Dodiya and Dr. K. B. Shukla are great fully acknowledged for their suggestions for the improvement of the manuscript. The authors gratefully acknowledge the financial assistance from the RKVY project “Validation of important crop varieties through DNA fingerprinting”.
REFRENCES
-
1Economic Survey. Ministry of Finance, Department of Economic Affairs.Economics Division Government of India, New Delhi. 2012-2013.
-
2Arumuganathan K and Earle ED . Nuclear DNA content of some important plant species. Plant Molecular Biology Report .1991; 9: 208-218.
-
3Saini M, Singh S, Hussain Z and Yadav A. RAPD analysis in mungbean (V. radiata (L.) Wilczek): I. Assessment of genetic diversity. Indian Journal of Biotechnolog. 2010;9: 137-146.
-
4Poehlman JM . The Mungbean. Oxford and IBH Publication, New Delhi. 1991.
-
5Kuleung C, Baenziger PS and Dweikar I. Transferability of SSR markers among wheat, rye, and triticale. Theoretical and Applied Genetics.2004; 108(6): 1147-1150.
-
6Xie RJ, Zhou J, Wang GY, Zhang SM, Chen L and Gao ZS. Cultivar identification and genetic diversity of Chinese bayberry (Myrica rubra) accessions based on fluorescent SSR markers. Plant Molecular Biology Reporter. 2011; 29(3): 554-562.
-
7Chen H, Qiao L, Wang L, Wanga L, Blair MW and Cheng Z. Assessment of genetic diversity and population structure of mung bean (Vigna radiata) germplasm using EST-based and genomic SSR markers. Gene . 2015; 566 (2): 175-183
-
8Chen H, Wang L, Wang S, Liu C, Blair MW and Cheng Z. Transcriptome Sequencing of Mung Bean (Vigna radiate L.) Genes and the Identification of EST-SSR Markers. PLoS ONE. 2015; 10(4): e0120273.
-
9Doyle JJ and Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochemical Bulletin.1987; 19: 11-15.
-
10Sambrook J, Fritschi EF and Maniatis T . Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York. 1989.
-
11Rohlf FJ. NTSYS-PC. Numerical taxonomy and multivariate analysis system.Exeter Software, New York. 1989.
-
12Sneath PHA and Sokal RR. Numerical Taxonomy. San Francisco, Freeman. 1973.
-
13Smith JSC, Chin EL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S and Zeigle J . An evaluation of the utility of SSR loci as molecular markers in maize (Z. mays L.):comparisons with data from RFLPs and pedigree. Theoretical and Applied Genetics.1997; 95: 163-173.
-
14Yeh FC, Boyle TJB. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian J. Bot.1997; 129:157-163.
-
15Singh SK, Lavanya GR, Bhat KV, Babu GS, Arya L, Verma M, Hussain Z, Roy S, Rathi RS and Mishra AK. Microsatellite Markers Revealed Genetic Diversity in Mungbean Mutant Lines. Indian Journal of Hill Farming. 2012; 25(1): 38-43.
-
16Gupta S, Gupta DS, Anjum KT, Pratap A and Kumar J. Transferability of simple sequence repeat markers in black gram (V. mungo L. Hepper). Australian Journal of Crop Science.2013; 7(3): 345-353.
-
17Sehrawat N, Bhat KV, Kaga A, Tomooka N, Yadav M and Jaiwal PK. Development of new gene-specific markers associated with salt tolerance for mungbean (Vigna radiata L.Wilczek). Spanish Journal of Agricultural Research. 2014;12 (3): 732-741.
-
18Gupta SK, Bansal R, Vaidya UJ and Gopalakrishna T. Assessment of genetic diversity at molecular level in mungbean (Vigna radiata (L.) Wilczek). Journal of Food Legumes. 2013; 26(3 & 4): 19-24.
-
19Dikshit HK, Jhang T, Singh NK, Koundal KR, Bansal KC, Chandra N, Tickoo JL and Sharma TR . Genetic differentiation of Vigna species by RAPD, URP and SSR markers. Biologia Plantarum. 2007; 51(3): 451-457.
-
20Liu C, Fan B, Cao Z, Su Q, Wang Y, Zhang Z, Wu J and Tian J. A deep sequencing analysis of transcriptomes and the development of EST-SSR markers in mungbean (Vigna radiata). Journal of Genetics. 2016; 95: 527-535.
-
21Goyal P, Jain R, Kachhwaha S and Kothari SL. Assessment of genetic diversity in Pithecellobium dulce (Roxb.) Benth. germplasm using RAPD and ISSR markers. Trees. 2015; 29: 637-653
Publication Dates
-
Publication in this collection
2018
History
-
Received
03 Feb 2016 -
Accepted
14 July 2016