ABSTRACT
The present study was concerned with the searching of novel bacterial cultures from different samples for the lab scale production of pectinase. Keeping in view the increasing demand of pectinase specially in Faisalabad, an industrial city of Pakistan, isolation of new hyper producer bacterial strains locally is an easy and cheap way of getting the desirable products at low cost. Therefore, isolation of new strains for industrial enzyme production has been and will be remained a part of research every time. This method alone can also provide raw material for further research such as enzyme engineering or molecular directed evolution. For the identification of hyper producer strain colony PCR was done for 16S rRNA analysis. Reason to use the 16S rRNA for identification purpose is that the gene is fairly short and can be amplified quickly and easily. The bacterial isolate (sources of pectinase enzyme) was identified based on PCR amplification of 16S rRNA and for this purpose the amplified product was run in agarose gel against a known species of Bacillus licheniformis. The 16S rRNA sequencing confirmed the Bacillus status of the strain.
Keywords:
indigenous; low cost; colony PCR; Bacillus licheniformis
INTRODUCTION
Enzymes are important biocatalysts used for various industrial and biotechnological purposes and produced by microorganisms, animals and plants. They can work in many adverse conditions compared to chemical catalysts. Microorganisms are minute creatures that can produce tremendous amounts of various metabolites and enzymes in very short span of time. Enzymes are usually produced from microorganisms such as yeast, fungi and bacteria but due to its rapid reproduction cycle, bacteria are considered as the most efficient bioproducers of enzymes. Microorganisms are considered as best source of enzyme production because of their short life, high yielding capacity and cost effectiveness. Approximately 50% of currently used enzymes are produced from fungi and yeast, 35% are produced from bacteria, while the remaining 15% are from plants and animals11 Barman S, Sit N, Badwaik LS, Deka SC. Pectinase production by Aspergillus niger using banana (Musa balbisiana) peel as substrate and its effect on clarification of banana juice. Journal of food science and technology. 2015;52(6):3579-89..
Great biodiversity of pectinolytic microorganisms are found in nature. Their common sources are soil22 Sunnotel O, Nigam P. Pectinolytic activity of bacteria isolated from soil and two fungal strains during submerged fermentation. World Journal of Microbiology and Biotechnology. 2002;18(9):835-9., rinds of fruits (peels), rotten vegetables and fruits33 Tariq A, Latif Z. Isolation and biochemical characterization of bacterial isolates producing different levels of polygalacturonases from various sources. African Journal of Microbiology Research. 2012;6(45):7259-64. and also their dump yards. Hence good pectinolytic bacteria can be obtained from samples of vegetables and fruit dumps or their peels may be used for isolation and screening. Bacteria are known to produce pectinases especially polygalacturonases44 Jayani RS, Shukla SK, Gupta R. Screening of bacterial strains for polygalacturonase activity: its production by Bacillus sphaericus (MTCC 7542). Enzyme research. 2010;2010.at different temperatures and pH55 Malvessi E, Silveira MMd. Influence of medium composition and pH on the production of polygalacturonases by Aspergillus oryzae. Brazilian Archives of Biology and technology. 2004;47(5):693-702.. As such many microbial enzymes are being used for biotechnological and industrial purposes. On the basis of pH requirement for their optimum activity, pectinases are classified into acidic and alkaline pectinase.
Acidic pectinases are useful in extraction, clarification and liquefaction of fruit juices66 Lee W, Yusof S, Hamid NSA, Baharin BS. Optimizing conditions for enzymatic clarification of banana juice using response surface methodology (RSM). Journal of Food Engineering. 2006;73(1):55-63. and wines77 Fernández M, Ubeda J, Briones A. Typing of non-Saccharomyces yeasts with enzymatic activities of interest in wine-making. International journal of food microbiology. 2000;59(1):29-36.,88 Cheirsilp B, Umsakul K. PROCESSING OF BANANA-BASED WINE PRODUCT USING PECTINASE AND a-AMYLASE. Journal of Food Process Engineering. 2008;31(1):78-90.. Whereas, alkaline pectinases are widely used in the fabric industry, pulp and paper industry99 Kohli P, Gupta R. Alkaline pectinases: A review. Biocatalysis and Agricultural Biotechnology. 2015;4(3):279-85.. Alkaline pectinases are among the most important enzymes and accounts for about 25% of worldwide sales of industrial enzymes1010 Chandel AK, Rudravaram R, Rao LV, Ravindra P, Narasu ML. Industrial enzymes in bioindustrial sector development: an Indian perspective. Journal of Commercial Biotechnology. 2007;13(4):283-91.. Pectinolysis at high pH and high temperature is potentially useful for application in the fabric industry, for the bio-preparation of cotton fabrics1111 Ahlawat S, Dhiman SS, Battan B, Mandhan R, Sharma J. Pectinase production by Bacillus subtilis and its potential application in biopreparation of cotton and micropoly fabric. Process Biochemistry. 2009;44(5):521-6. and enzymatic polishing of cotton blended fabrics1212 Mccloskey SG, Jump JM. Bio-polishing of polyester and polyester/cotton fabric. Textile research journal. 2005;75(6):480-4. and in the paper industry1313 Ahlawat S, Mandhan R, Dhiman SS, Kumar R, Sharma J. Potential application of alkaline pectinase from Bacillus subtilis SS in pulp and paper industry. Applied biochemistry and biotechnology. 2008;149(3):287-93..
Due to its applicability in various fields of biotechnology and industry, the demand for commercial pectinase with novel characteristics and high stability is increasing. Pectinase constitute about 10% from overall enzymes used in various industrial processes. Large number of pectinase producing microorganisms has already been explored and characterized, However, as demand for pectinase is increasing, further exploration of new sources of pectinase with different characters is actually become the need of the day.
Researchers nowadays have focused their attention towards the exploration of new microbial isolates with desirable biochemical and physicochemical characteristics and a low cost production1414 Ghazala I, Haddar A, Romdhane MB, Ellouz-Chaanouni S. Screening and Molecular Identification of New Microbial Strains for Production of Enzymes of Biotechnological Interest. Brazilian Archives of Biology and Technology. 2016;59.,1515 Karthik L, Kumar G, Rao KB. Screening of pectinase producing microorganisms from agricultural waste dump soil. Asian Journal of Biochemical and Pharmaceutical Research. 2011;1(2):329-337..There are many reports on pectinolytic microorganism isolated from different geographical locations and sources especially from agro/industrial wastes and spoiled fruits1616 Geetha M, Saranraj P, Mahalakshmi S, Reetha D. Screening of pectinase producing bacteria and fungi for its pectinolytic activity using fruit wastes. International journal of biochemistry & biotech science. 2012;1:30-42.,33 Tariq A, Latif Z. Isolation and biochemical characterization of bacterial isolates producing different levels of polygalacturonases from various sources. African Journal of Microbiology Research. 2012;6(45):7259-64..
Enzymes particularly, due to their specific nature of activity, are widely utilized in various processes in different sectors of industrial, environmental and food biotechnology. In order to extract pectinase, microbes are the best source as they allow an economical technology with low resource consumption. Enzymes may be obtained by screening microorganisms sampled from new localities or developed by alteration of the present enzymes using modern methods of protein engineering. Due to increasing demand of enzymes in global market, new enzymes for commercial application with desirable biochemical and physico-chemical characteristics and a low cost production have become a focus of research and finding enzyme producing microorganisms are a major subject of research. New microorganisms capable of producing huge amounts of enzymes of interest with different characteristics from cost effectiverenewable resources are a need of the day because this alone can be a way to increase enzyme production without using any costly materials.
The present study was concerned with the searching of novel bacterial cultures from different soil samples for the lab scale production of pectinase. The bacterial isolates (sources of pectinase enzyme) were identified based on PCR amplification of 16S rRNA.
MATERIALS AND METHODS
Isolation of Pectinolytic Bacterial Strain
Pectinolytic bacterial strains were isolated from different natural habitats such as soil, textile wastes, garden compost, rotten fruits and vegetables.
Isolation was carried out by the following method:
-
Collection of sample was done from various sources i.e., soil, garden compost, rotten fruits and vegetables. The samples were collected in polythene bags and were kept in cold cabinet till further investigation. After 24hours, 500µl of the above overnight culture was poured and spread on Pectinase screening agar medium (PSAM) petri plates.
-
About 10g of each sample was stirred in 50ml autoclaved distilled water in separate beakers and about 2ml of each sample was enriched in 100ml autoclaved nutrient medium.
-
Pectinase Screening Agar Medium (PSAM) was prepared containing the following composition (g/L; NaNO3 2g, KCl 0.5g, MgSO4 0.5g, K2HPO4 1g, trypton 0.5g, agar 20g and pectin 10g). PSAM was poured in petri plates and streaked by the culture of above medium. The Petri plates were incubated at 37°C for 24hrs and checked for clear zone formation.
-
The hyper producer colonies were identified by treating the plates with potassium iodide solution and re-streaked on the same agar medium to maintain a pure culture of the pectinolytic bacterial strain for further study.
Maintenance of Isolates
The desired isolate was cultured on pectin yeast extract agar medium of the following composition (g/L; NaNO3 2g, KCl 0.5g, MgSO4 0.5g, KH2PO4 1g, yeast extract 1g, agar 20g and pectin 5g. The Petri plates were incubated at 37°C for two days and then stored in a refrigerator at 4°C for culturemaintenance. Glycerol stock of the pure culture was preserved for further use.
Submerged Fermentation
A single colony from pure culture was picked and inoculated in LB medium. 100µl of the above overnight culture was sub-cultured in a medium with different pectin concentration. Media prepared has the following basic W/V composition: NaNO3; 0.2%, KH2PO4; 0.1%, KCl; 0.05%, MgSO4; 0.05% and trypton; 0.5% in addition to variable quantity of pectin. 100ml media was prepared in 250ml flask and sterilized at 121°C for 20 min in autoclave. The prepared medium was cooled to room temperature and inoculated by 0.3mg (on dry cell mass basis) concentration ofinoculums of the selected strain, transferred to 100ml medium. The inoculum was prepared fresh as above. For every experiment, after completion of required incubation period, medium was centrifuged at 4°C and 5432xg for 10 min to obtain clear supernatant. Supernatant was used for enzyme activity assay.
Pectinase Assay
One unit of pectinase activity is defined as the number of reducing sugar (galacturonic acid) obtained from 1ml of pectinase in 1 min. The resulted galacturonic acid was analyzed using modified1717 Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical chemistry. 1959;31(3):426-8. method. Measurement of pectinase activity was carried out using 0.7ml of 0.5% pectin dissolved in 0.1M Glycine buffer pH 8 and 0.3ml of crude enzyme. This reaction mixture was incubated at 70°C for 10 min in shaking water bath. 1ml of DiNitro Salicylic Acid (DNS) was added to the mixture and boiled in water bath 100°C for 10 min. The OD of the mixture was measured at 575 nm absorbance using UV-VIS spectrophotometer.
Identification of Isolated Strains
Identification of selected pectinolytic bacterial strain was carried out by PCR based 16S rRNA amplification and subsequent sequencing of the product to identify the bacteria to the species level.
RESULTS AND DISCUSSION
Isolation of Bacteria
Keeping in view the increasing demand of enzymes in various industrial applications, isolation of new hyper producer bacterial strains for over production of desirable metabolites is an easy and cheap way of getting the desirable products locally at low cost and therefore isolation of new strains for industrial enzyme production has been and will be remained a part of research every time. This method alone can provide raw material for further research based on modern technologies of producing new enzymes with different characters such as enzyme engineering or molecular directed evolution.
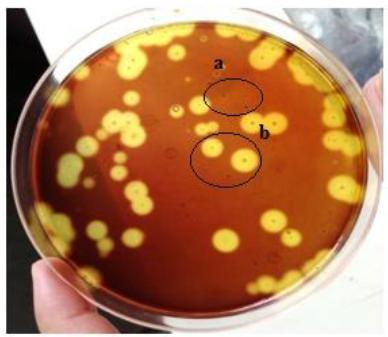
The bacteria from different samples were enriched in nutrient broth and cultured on petri plates by spreading method and after incubation of 24hrs at 37°C, plates were washed with potassium iodide solution. As is clear from Figure 1, pectinolytic bacteria showed clear zone when treated with potassium Iodide solution while those bacteria that had no pectinolytic activity did not form any clear zone after potassium Iodide solution treatment.
Bacterial isolation from sample (PSAM petri plate showing mixed culture). Colonies appear as brown dots on agar plate. (a) Colonies without pectinolytic activity, (b) Colonies with pectinolytic activity and clear zone formation.
The hyper producer colonies as shown in Figure 2 A and B, having largest clear zones were identified and streaked on the same agar medium to maintain a pure culture of the desired pectinolyticbacterial strain for further study.The control plate (C) with no pectinolytic bacterial colony has no clear zone. Isolation of pectinase producing bacteria has been carried out by many researchers1818 Mikicinski A, Pulawska J, Sobiczewski P, Orlikowski L. Pectolytic bacteria associated with soft rot of dieffenbachia (Dieffenbachia maculata). Phytopathologia. 2010;58: 21-32.
19 Zeni J, Cence K, Grando CE, Tiggermann L, Colet R, Lerin LA, et al. Screening of pectinase-producing microorganisms with polygalacturonase activity. Applied biochemistry and biotechnology. 2011;163(3):383-92.-2020 Silva D, Martins EdS, Silva Rd, Gomes E. Pectinase production by Penicillium viridicatum RFC3 by solid state fermentation using agricultural wastes and agro-industrial by-products. Brazilian Journal of Microbiology. 2002;33(4):318-24. from different and diverse sources such as soil, rotten fruits and vegetables and their dump yards. Keeping in view the textile industry of the city (Faisalabad, Pakistan), alkaline pectinase producing bacterial strains were isolated and checked for their pectinolytic activity.
(A) Hyper-producer colony with largest clear zone, (B) Hyper producer colony isolated and re-grow on other plate, (C) Control with no colony and no clear zone.
Plate Assay Method for Pectinase Qualitative assay
As shown in Figure 2 A, B and C, the control did not show any clear zone while the well defined clear zones are visible around colonies having pectinase activity. The diameter of the clear zone is directly proportional to the pectinase activity. It means that colonies with large clear zones possess high pectinase quantity while those with low diameter clear zone have low activity. This is also a simple activity assay. Such an assay method remained an identification tool for specific enzyme detection and used by many researchers2121 McKay A. A plate assay method for the detection of fungal polygalacturonase secretion. FEMS Microbiology letters. 1988;56(3):355-8.
22 Soares MM, Silva Rd, Gomes E. Screening of bacterial strains for pectinolytic activity: characterization of the polygalacturonase produced by Bacillus sp. Revista de Microbiologia. 1999;30(4):299-303.-2323 Oliveira K, Malavolta L, Souza C, Vicente E, Laluce C. Pectinolytic activity secreted by yeasts isolated from fermented citrus molasses. Journal of applied microbiology. 2006;100(4):633-40..
Submerged fermentation
In submerged fermentation, the selected strain produced 39U/ml with different concentrations of pectin. Submerged fermentation of bacterial cultures produce various metabolites in short time and can be manipulated easily by addition of specific substrates to produce desirable metabolites. The production can be increased easily by changing medium physical conditions such as pH, temperature and agitation rate or its chemical composition55 Malvessi E, Silveira MMd. Influence of medium composition and pH on the production of polygalacturonases by Aspergillus oryzae. Brazilian Archives of Biology and technology. 2004;47(5):693-702..
Identification of isolated bacterial strain
The hyper producer bacterial strain was streaked on fresh petri plate of PSAM and a single colony was picked and colony PCR was done.
The hyper producer strain was selected for 16S rRNA analysis and for this purpose colony PCR was done and the amplified product was run in agarose gel against a known species of Bacillus licheniformis and as is revealed from Figure 3, both the bands (a) and (b) appeared on the same level in the agarose gel thus indicating the similarity of newly isolated bacterial strain with Bacillus licheniformis. Later on 16S rRNA sequencing confirmed the Bacillus status of the strain. The 16S rRNA sequence was submitted to National Center for Biotechnology Information (NCBI)GenBank and accession No. allotted by it for the newly isolated Bacillus licheniformis strain is “KX765286”. Colony PCR is an efficient way for identification of species2424 Mirhendi H, Diba K, Rezaei A, Jalalizand N, Hosseinpur L, Khodadadi H. Colony PCR is a rapid and sensitive method for DNA amplification in yeasts. Iranian Journal of Public Health. 2007;36(1):40-4.,2525 Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. Journal of Clinical Microbiology. 2002;40(12):4744-7..
Gel Electrophoresis of Colony PCR product to reveal 16S rRNA. a). 16S rRNA of newly isolated Bacteria, b). Control (16S rRNAof known species of Bacillus licheniformis).
All bacteria possess a gene for the 16S rRNA located on 30S ribosomal unit, the DNA sequence of which is species specific. So it possible to use the sequence of this gene to identify a bacterial species, rather than guessing based on observed characteristics. Another reason to use the 16S rRNA for identification purposes is that the gene is about 1500 base pairs. It is a short gene and can be amplified easily and quickly.
CONCLUSIONS
Bacteria especially the genus Bacillus represents the most efficient group of pectinolytic microorganisms. Isolation of new and local strains is a prerequisite for development and production of enzymes with novel and desirable characteristics. Colony PCR is a rapid and reliable method for identification of microorganisms. PSAM is also a rapid method of detecting pectinolytic activity and helps in identification and selection of pectinase producing bacteria. Thus with the help of these tools a researcher can add new and novel species to obtain their desirable metabolites to be used for various purposes.
ACKNOWLEDGMENTS
Will be included later on after final decision about manuscript.
REFERENCES
-
1Barman S, Sit N, Badwaik LS, Deka SC. Pectinase production by Aspergillus niger using banana (Musa balbisiana) peel as substrate and its effect on clarification of banana juice. Journal of food science and technology. 2015;52(6):3579-89.
-
2Sunnotel O, Nigam P. Pectinolytic activity of bacteria isolated from soil and two fungal strains during submerged fermentation. World Journal of Microbiology and Biotechnology. 2002;18(9):835-9.
-
3Tariq A, Latif Z. Isolation and biochemical characterization of bacterial isolates producing different levels of polygalacturonases from various sources. African Journal of Microbiology Research. 2012;6(45):7259-64.
-
4Jayani RS, Shukla SK, Gupta R. Screening of bacterial strains for polygalacturonase activity: its production by Bacillus sphaericus (MTCC 7542). Enzyme research. 2010;2010.
-
5Malvessi E, Silveira MMd. Influence of medium composition and pH on the production of polygalacturonases by Aspergillus oryzae. Brazilian Archives of Biology and technology. 2004;47(5):693-702.
-
6Lee W, Yusof S, Hamid NSA, Baharin BS. Optimizing conditions for enzymatic clarification of banana juice using response surface methodology (RSM). Journal of Food Engineering. 2006;73(1):55-63.
-
7Fernández M, Ubeda J, Briones A. Typing of non-Saccharomyces yeasts with enzymatic activities of interest in wine-making. International journal of food microbiology. 2000;59(1):29-36.
-
8Cheirsilp B, Umsakul K. PROCESSING OF BANANA-BASED WINE PRODUCT USING PECTINASE AND a-AMYLASE. Journal of Food Process Engineering. 2008;31(1):78-90.
-
9Kohli P, Gupta R. Alkaline pectinases: A review. Biocatalysis and Agricultural Biotechnology. 2015;4(3):279-85.
-
10Chandel AK, Rudravaram R, Rao LV, Ravindra P, Narasu ML. Industrial enzymes in bioindustrial sector development: an Indian perspective. Journal of Commercial Biotechnology. 2007;13(4):283-91.
-
11Ahlawat S, Dhiman SS, Battan B, Mandhan R, Sharma J. Pectinase production by Bacillus subtilis and its potential application in biopreparation of cotton and micropoly fabric. Process Biochemistry. 2009;44(5):521-6.
-
12Mccloskey SG, Jump JM. Bio-polishing of polyester and polyester/cotton fabric. Textile research journal. 2005;75(6):480-4.
-
13Ahlawat S, Mandhan R, Dhiman SS, Kumar R, Sharma J. Potential application of alkaline pectinase from Bacillus subtilis SS in pulp and paper industry. Applied biochemistry and biotechnology. 2008;149(3):287-93.
-
14Ghazala I, Haddar A, Romdhane MB, Ellouz-Chaanouni S. Screening and Molecular Identification of New Microbial Strains for Production of Enzymes of Biotechnological Interest. Brazilian Archives of Biology and Technology. 2016;59.
-
15Karthik L, Kumar G, Rao KB. Screening of pectinase producing microorganisms from agricultural waste dump soil. Asian Journal of Biochemical and Pharmaceutical Research. 2011;1(2):329-337.
-
16Geetha M, Saranraj P, Mahalakshmi S, Reetha D. Screening of pectinase producing bacteria and fungi for its pectinolytic activity using fruit wastes. International journal of biochemistry & biotech science. 2012;1:30-42.
-
17Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical chemistry. 1959;31(3):426-8.
-
18Mikicinski A, Pulawska J, Sobiczewski P, Orlikowski L. Pectolytic bacteria associated with soft rot of dieffenbachia (Dieffenbachia maculata). Phytopathologia. 2010;58: 21-32.
-
19Zeni J, Cence K, Grando CE, Tiggermann L, Colet R, Lerin LA, et al. Screening of pectinase-producing microorganisms with polygalacturonase activity. Applied biochemistry and biotechnology. 2011;163(3):383-92.
-
20Silva D, Martins EdS, Silva Rd, Gomes E. Pectinase production by Penicillium viridicatum RFC3 by solid state fermentation using agricultural wastes and agro-industrial by-products. Brazilian Journal of Microbiology. 2002;33(4):318-24.
-
21McKay A. A plate assay method for the detection of fungal polygalacturonase secretion. FEMS Microbiology letters. 1988;56(3):355-8.
-
22Soares MM, Silva Rd, Gomes E. Screening of bacterial strains for pectinolytic activity: characterization of the polygalacturonase produced by Bacillus sp. Revista de Microbiologia. 1999;30(4):299-303.
-
23Oliveira K, Malavolta L, Souza C, Vicente E, Laluce C. Pectinolytic activity secreted by yeasts isolated from fermented citrus molasses. Journal of applied microbiology. 2006;100(4):633-40.
-
24Mirhendi H, Diba K, Rezaei A, Jalalizand N, Hosseinpur L, Khodadadi H. Colony PCR is a rapid and sensitive method for DNA amplification in yeasts. Iranian Journal of Public Health. 2007;36(1):40-4.
-
25Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. Journal of Clinical Microbiology. 2002;40(12):4744-7.
Publication Dates
-
Publication in this collection
2018
History
-
Received
27 July 2016 -
Accepted
14 Sept 2016