ABSTRACT
LOB (lateral organ boundaries)-domain proteins define a family of plant-specific transcription factors involved in developmental process from embryogenesis to seed production. They play a crucial role in shaping the plant architecture through coordinating cell fate at meristem to organ boundaries. Identification of LBD genes from Brassica rapa genome, and analysis of phylogeny,gene structure, chromosome location, phylogenetic and tissue expression pattern analysis of LBD family genes in Chinese cabbage will be useful to the functions identification of plant LBD genes. Based on Brassica rapa genome database and bioinformatic method, Chinese cabbage LBD family genes were identified and the genes were sequenced. A phylogenetic tree was created using the MEGA5 program. Gene structure and chromosomes location were done by MapDraw, GSDS and Clustal X. Expression pattern of LBD genes at different development stages was analyzed based on RNA-seq. A total of 62 LBD genes were identified and could be classified into two classes and four subclasses according to the gene structure and conserved domain phylogeny relationship. Distribution mapping showed that the predicted LBDs were unevenly localized on all the 10 chromosomes, suggesting that they have an extensive distribution on the Brassica rapa chromosomes. Most of the LBD genes had differential expression pattern and showed highly diverse tissue-specific expression and functional diversity. To our knowledge, this is the first report of a genome wide analysis of the Brassica rapa LBD gene family, which would provide valuable information for understanding the classification and putative functions of the gene family.
Keywords:
Brassica rapa; LBD; phylogenetic analysis; gene expression; genome-wide
INTRODUCTION
LBD (LATERAL ORGAN BOUNDARIES domain) gene family is an important transcript factor family that has been assigned to this functional group on the basis of its nuclear localization and capacity to bind to a DNA motif 11 Husbands A, Bell EM, Shuai B, Smith HMS, Springer S. LATERAL ORGAN BOUNDARIES: defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007; 35(19): 6663-6671.
2 Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009; 151(3): 1377-1389.-33 Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011; 16(1): 47-52.. Until now, LBD genes were found only in plant databases indicating that this unique gene family may only regulate plant-specific processes 44 Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel plant-specific gene family. Plant Physiol. 2002; 129(2): 747-761.. The LBD gene family can be divided into two classes according to the structure of the LOB domain in the N terminus. Class I LBD genes contain a perfectly conserved CX2CX6CX3C zinc finger-like domain with a length of about 100 amino acids which is presumably required for DNA binding; while class II LBD genes have only an LX6LX3LX6L leucine zipper-like coiled-coil motif, which is probably involved in protein dimerization 33 Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011; 16(1): 47-52.,55 Landschaslz WH, Johnson PF, McKnight SL. The lucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1998; 240(4860): 1759-1765.. The LOB structure often also contains a GAS domain with a 49 amino acid sequence, which generally begins with the FX2VH sequence and ends with a DP (V/I) YG sequence. The number of amino acids in the C-domain of the two types of LBD genes was consistent, but the amino acid species was different. The consensus sequence of class I LBD gene is CAACKFLRRKCX3C, while that of class II LBD gene is CNGCRVLRKGCSE (D/N) C 44 Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel plant-specific gene family. Plant Physiol. 2002; 129(2): 747-761.,66 Semiarti E, Ueno Y, Tsukay H, Iwakawa H, Machida C, Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001; 128(10): 1771-1783.,77 Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, et al. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007; 51(2): 173-184.. LBD proteins have varied expression patterns ranging from temporal to tissue differences, suggesting that they may function in diverse processes. Numerous LBD genes are expressed at the adaxial base of plant lateral organs, they genes play critical roles in lateral organ development during a plant’s growth 88 Sun SB, Song JP, Meng LS. ASYMMETRIC LEAVES2 gene a member of LOB/AS2 family of Arabidopsis thaliana causes an abaxializing leaves in transgenic cockscomb. Mol Biol Rep. 2012; 39(4): 4927-4935.-99 Chalfun-Junior A, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family controls proximal-distal patterning in Arabidopsis petals. Plant Mol Biol. 2005; 57(4): 559-575..
To date, a variety of LBD gene family have been successfully identified and investigated in some plants, including Arabidopsis thaliana, rice, poplar, tomato, Malus, Medicago truncatula, maize, pepper, Nicotiana tobacco and Brachypodium, which contains 43, 35, 57, 46, 58, 56, 44, 45, 98 and 28 LBD genes, respectively 44 Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel plant-specific gene family. Plant Physiol. 2002; 129(2): 747-761.,1010 Yang Y,Yu X,Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006; 39(1): 248-262.
11 Zhu QH,Guo AY,Gao G,Zhong YF,Xu M,Huang M,et al. DPTF:a database of poplar transcription factors. Bioinformatics. 2007; 23(10): 1307-1308.
12 Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana and functional and molecular comparisons between AS2 and other family members. Plant J. 2009; 58(3): 525-537.
13 Wang XF, Liu X, Su L, Sun YJ, Zhang S, Hao YJ, et al. Identification evolution and expression analysis of the LBD gene family in tomato. Scientia Agri Sin. 2013; 46(12): 2501-2513.
14 Wang X, Zhang S, Su L, Liu X, Hao Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. Plos One. 2013; 8(2): e57044.
15 Jia XT, Liu WX, Xie WG, Liu ZP, Liu ZM, Wang YR. Genome-wide analysis of the LBD transcription factor family in Medicago truncatula. Acta Bot Boreal-Occident Sin. 2014; 34(10): 2176-2187.
16 Zhang YM,Zhang SZ,Zheng CC. Genome wide analysis of LATERAL ORGAN BOUNDARIES domain gene family in Zea mays. J Genet. 2014; 93(1): 79-91.
17 Zheng ZF, Zhang YL, H C, Dai XZ, Liu F, Yuan ZH. Genome-wide Identification and Expressing Analysis of LBD Transcription Factors in Pepper. Acta Hortic Sin. 2016; 43(1): 683-694.
18 Sun TT, Gong DP, Zhang L, Chen YQ, Zhao W, Xiang XH, et al. Genome-wide Sequence Identification and Expression Analysis of the LBD Gene Family in Nicotiana tabacum. J Plant Genet Resour. 2016; 17(2): 316-325.-1919 Gombos M, Zombori Z, Szécsényi M, Gyorgyey J. Characterization of the LBD gene family in Brachypodium: a phylogenetic and transcriptional study. Plant Cell Rep. 2017; 36(1): 61-79.. What is more, several members of the LBD family have been functionally identified in different species. In Arabidopsis, AtLBD04 is involved in regulating leaf development 2020 Dolan L, Langdale JA. New insights into plant development in New England. Development. 2004; 131(21): 5215-5220.; AtAS2 is specially expressed in young floral organs, which regulates floral organ development 77 Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, et al. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007; 51(2): 173-184.; AtLBD16, AtLBD17, AtLBD18 and AtLBD29 can combine auxin signaling pathway with other cell processes, then regulates the regeneration of lateral roots and callus formation 2121 Fan M, Xu C, Xu K, Hu YX. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012; 22(7): 1169-1180.-2222 Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, et al. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell. 2014; 26(3): 1081-1093.; AtLBD37, AtLBD38 and AtLBD39 that induced by nitrate are involved in anthocyanin synthesis and nitrate metabolism 2323 Rubin G, Tohge T, Matsuda F, Saito K, Scheible W. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009; 21(11): 3567-3584.-2424 Albinsky D, Kusano M, Higuchi M, Hayashi N, Kobayashi M, Fukushima A, et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol Plant. 2010; 3(1): 125-142.; gibberellin inhibits the expression of AtASL37 2525 Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007; 19(11): 3037-3057.,in contrast, AtAS2 is able to promote gibberellin synthesis 2626 Ikezaki M, Kojima M, Sakakibara H, Kojima S, Ueno Y, Machida C, et al. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 2010; 61(1): 70-82.. In poplar, PtaLBD1 regulates secondary growth, while PtaLBD15 and PtaLBD18 are specifically expressed in secondary xylem, suggesting that the LBD family is involved in secondary growth during xylem formation 2727 Yordanov YS,Regan S,Busov V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell. 2010; 22(11): 3662-3677.. The maize ZmLBD19 gene dimerize with the maize AtAS1 ortholog RS2 is a key regulator of female gametophyte development and leaf axial differentiation 2828 Evans MMS. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell. 2007; 19(1): 46-62.. The Rice OsIG1 (homologous to Arabidopsis
AS2) can influence the lateral growth of leaves by regulating the division of follicular cells between vascular bundles 2929 Zhang J,Tang W,Huang Y,Niu X,Zhao Y,Han Y,et al. Down-regulation of a LBD-like gene,OsIG1,leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J Exp Bot. 2015; 66(1): 99-112..
The crops of the genus Brassica are mainly used as vegetables, oilseed and fodder. Their yields in China account for more than half of that in the world and 61% of that in Asia (http://faostat.fao.org). Brassica rapa is one of the most important vegetables in China and is cultivated extensively worldwide. Given its significant economic value and close relationship to Arabidopsis, the Brassica rapa (Chiifu-401-42) genome was sequenced and assembled 3030 Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature genet. 2011; 43(10): 1035-1039..
However, there was no report about the LBD gene family of Brassica rapa, despite the important role of these proteins in plant growth and development. Therefore, an investigation of the LBD genes in the whole genome of Brassica rapa is timely.
In this study, we comprehensively described BrLBD genes by comparative genomic analysis. The aims of this study were as follows: (1) to identify and map BrLBD genes onto ten chromosomes; (2) to classify BrLBD genes through comparative genomics analysis; (3) to identify orthologous and paralogous LBD genes; (4) to analyze BrLBD expression patterns in five tissues using RNA-seq. This study provides useful resources for future studies on the structure and function of BrLBD genes, as well as for identifying and characterizing LBD genes in other species.
MATERIALS AND METHODS
Identification of LBD genes in Brassica rapa
To identify the members of the LBD gene family in Brassica rapa (Chiifu-401-42), the following strategy was performed. First, the Brassica rapa genome sequence is known and filtered protein and CDS sequences are available 3030 Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature genet. 2011; 43(10): 1035-1039.. Whole genome proteins of two species were downloaded, including Brassica rapa (http://brassicadb.org/brad-/geneFamily.php) and Arabidopsis (http://datf.cbi.pku.edu.cn/). All annotated LBD members in the Brassica rapa genome database were selected. Second, we analyzed the typical domain of LBD (DUF260, PF03195) using a hidden Markov model (HMM) 3131 Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998; 23(10): 403-405.-3232 Wu X, Song C, Wang B, Cheng J. Hidden Markov model used in protein sequence analysis. J Biomed Eng. 2002; 19(1): 455-458. analysis with Pfam searching, from the Brassica rapa genome sequences using a Perl-based script. Then, all of the protein sequences derived from the collected candidate LBD genes were examined using the domain analysis programs Pfam (http://pfam.sanger.ac.uk/) and SMART (http://smart.emblheidelberg.de/) 3333 Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012; 40(D1): 302-305. with the default cutoff parameters, and repetitive LBD genes were removed manually. Finally, all candidate LBD genes meeting these standards were compared with known AS2/LOB domain sequences using ClustalX (http://www.clustal.org/) to eliminate the redundancy sequences that not containing the signature conserved domain of LBDs 3434 Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004; 32(suppl 1): D138-D141.. The isoelectric point (pI) and molecular weight (MW) were computed using Expasy tools (http://web.expasy.org/compute_pi/) 3535 Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012; 40(W1): W597-W603.. The chromosomal locations and the exon/intron information were obtained from the Phytozome database 3636 Goodstein DM, Shu S, Howson R, Hayes RD, Fazo J, Mitros T, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40(D1): 1178-1186. using a Perl-based program.
Chromosomal location and LBD genes structure analysis
The chromosomal locations were retrieved from the genome data downloaded from the Phytozome database 3636 Goodstein DM, Shu S, Howson R, Hayes RD, Fazo J, Mitros T, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40(D1): 1178-1186. using a Perl-based program and mapped to the chromosomes using the MapDraw tools 3737 Liu RH, Meng JL. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas. 2003; 25(3): 317-321. as well as the gene structure of the LBD genes were generated with the GSDS (http://gsds.cbi.pku.edu.cn/) 3838 Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Hereditas. 2007; 29(8): 1023-1026..
Sequence alignment and phylogenetic analysis
To identify signature domains, Clustal X (version 1.83) was used to align amino acid sequences of LBD proteins. To understand the evolutionary relationships between the Brassica rapa LBD proteins and the variations in LBD sequences, AtLBDs and BrLBDs were selected for phylogenetic tree analysis using MEGA5 (http://www.megasoftware.net/) 3939 Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011; 28(10): 2731-2739.. Initially, the retrieved Brassica rapa and Arabidopsis LBD nucleotide sequences were translated into amino acid sequences using BioXM 2.6 in the fasta format, and protein sequences were then aligned using Clustal X 4040 Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31(13): 3497-3500.. MEGA analysis was conducted after these steps. The Maximum Likelihood (ML) method was performed with the complete deletion option. For statistical reliability, bootstrap analysis was conducted with 1000 replicates to assess statistical support for each mode.
LBD genes expression
Brassica rapa tissue expression information from raw RNA-seq data were downloaded from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE43245 4141 Tong C, Wang X, Yu J, Wu J, Li W, Huang J, et al. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics. 2013; 14(1): 689.. The mRNAs of these transcriptome data were extracted from six tissues (root, stem, leaf, flower, and silique) of Chiifu-401-42. Expression profile cluster analysis of the Brassica rapa LBD family proteins was constructed using MEV Software 4242 Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free open-source system for microarray data management and analysis. Biotechniques. 2003; 34(2): 374-378..
RESULTS
Identification and annotation of the LBD genes in Brassica rapa
To identify the LBD proteins in Brassica rapa, a local BLAST program and the HMM of the SMART and Pfam tools were used, and a total of 62 LBD genes from the entire Brassica rapa genome were identified (Table 1). According to multiple sequence alignment results of the LBD domain, 53 and 9 LBD genes were identified as class I and class II, respectively. The result indicates that most LBD genes in Brassica rapa belong to the class I. In addition, we analyzed the gene identifier, genomic position, length of coding sequence, as well as length of amino acid sequence, pI, and MW of these LBD genes. The length of LBD proteins ranged from 131 (Bra039072) to 605 (Bra021627) amino acids (aa), the pIs of the LBD proteins were between 4.72 (Bra019364) and 9.68 (Bra030647), with a distribution from acidic to alkaline; the MWs of these proteins were between 14.56 kDa (Bra025294) and 66.85 kDa (Bra032430).
Phylogenetic and structure analysis of the LBD genes in Brassica rapa
To evaluate the evolutionary relationships among the 62 BrLBD proteins, we performed a phylogenetic analysis based on their full-length amino acid sequences. We identified two subfamilies (class I and class II) as being monophyletic (Fig. 1) including 53 and 9 LBD genes. And 23 paralogous LBDs were found, 18 of which had a very strong bootstrap support (>90%). Our results suggest a clear paralogous pattern of LBD gene divergence by gene duplication for the Brassica rapa.
The phylogenetic tree and gene structure analysis of the LBD gene family in Brassica rapa. The amino acid sequences of the LBD proteins were aligned with ClustalX, and the phylogenetic tree was constructed using the neighbor joining method in MEGA5 software. Each node is represented by a number that indicates the bootstrap value for 1000 replicates. The right side illustrates the exon-intron organization of the corresponding LBD genes. The exon and intron are represented by the yellow boxes and black lines, respectively. The scale bar represents 1kb (right).
Structural analyses were intended to provide valuable information concerning duplication events when interpreting phylogenetic relationships within gene families. Thus, we analyzed the exon/intron structures of the LBD genes (Fig. 1). In Brassica rapa, the exon numbers ranged from one within 14 genes to eight in Bra32430. 39 genes had two exon, six genes had three exons, and two gene each contain five (Bra032938) and six exons (Bra021627). Interestingly, eight genes of class II all contain only two exon. Most members within the same subgroup shared a similar intron/exon structure and gene length. The conserved intron/exon structure in each subgroup supported their close evolutionary relationships and the stated classification of subfamilies.
To compare the evolutionary patterns of Brassica rapa LBD gene family with those of other plants, a phylogenetic tree was generated using Brassica rapa, Arabidopsis, tomato and maize full length protein sequences (Fig. 2). The results show that the class I BrLBD genes could be subdivided into four subgroups of Ia, Ib, Ic and Id, which contained 9, 15, 12, 17 LBD family members, respectively, which indicated that the LBD family of Brassica rapa were distributed each subclass. We identified 29 pairs of orthologous genes among all LBD genes. Conversely, 22 orthologous gene pairs were found between Brassica rapa and Arabidopsis, 3 orthologous gene pairs between Brassica rapa and tomato, two orthologous gene pairs between tomato and maize, and two orthologous gene pairs between tomato and maize. However, we did not detect orthologous gene between Brassica rapa and maize, which was consistent with the close relationship between Brassica rapa and Arabidopsis. In addition, many paralogous LBD gene pairs were identified in maize (11), tomato (8), Brassica rapa (4) and Arabidopsis (3).
Phylogenetic tree constructed using the Maximum Likelihood method by MEGA5, using LBD genes in Brassica rapa, Arabidopsis, tomato and maize. Branches of members belonging to class II subclasses are represented by black lines, branches of members belonging to class Ia, class Ib , class Ic and class Id subclasses are represented by red, blue, green and yellow lines, respectively. The black hollow circles represent paralogous genes of Brassica rapa, and the black solid circles represent orthologous genes from Brassica rapa and Arabidopsis.
Chromosomal localization of Brassica rapa LBD genes
Chromosomal location analyses showed that 62 BrLBD genes presented on 10 chromosomes by the MapDraw tool, which dispersed throughout their respective genomes (Fig. 3). The number of LBD genes on each chromosome varied widely. The largest number of LBD genes was detected on chromosome A04 and A09 (11 BrLBDs), while the lowest number was on chromosomes A08 and A10 (one BrLBDs each). Chromosomes A01, A06, and A07 had the same number of LBD genes (5 BrLBDs). Eight and six LBD genes were located on chromosome A05 and A02, respectively. Interestingly, a pair paralogous LBDs (Bra019365 and Bra019364) were located in the same chromosome (A03) and all other pairs located on different chromosomes. Further investigation showed that three pairs of BrLBDs (Bra004572 and Bra004693, Bra004908 and Bra004910, Bra018260 and Bra018320) closely linked in chromosome A05. In addition, some members were clustered together at the top of chromosome A09, including Bra037322, Bra037323, Bra0390372, Bra035860, Bra037847 and Bra037142.
The chromosomal mapping analysis of the LBD gene family in Brassica rapa. The chromosome number (Chr01-Chr10) is indicated at the top of each chromosome.
Sequence alignment and conserved motifs of LBD genes
In Arabidopsis, the LBD had a conserved AS2/LOB domain in the N terminus of the proteins, and there were two conserved blocks in the AS2/LOB domain of the class I proteins, i.e., the C block and GAS block. To identify conserved domains within the BrLBDs, we performed an alignment within all the LBD genes and a separate one for the class I and II protein sequences. As with the LBD genes, multiple sequence alignments showed that all 62 predicted LBD protein sequences had a high conserved CX2CX6CX6C zinc-finger-like domain while a LX6LX3LX6L leucine-zipper-like domain existed only in 53 genes of the class I (Fig. 4). However, two points are worth mentioning: three genes (Bra019363, Bra019364 and Bra019365) had seven amino acid residues between the third and fourth cysteine (C), which differ from other genes had three. Bra018260 had an insertion with 13 amino acid residues between the first and second leucine (L).
Multiple sequence alignment of amino acid sequences of conserved domain motif of BrLBD proteins. A: The CX2CX6CX3C zinc finger-like domain sequences Logos. B: The LX6LX3LX6L Leu-zipper-like domain sequences Logos. Sequence alignment of two domains by ClustalX and conserved motifs Logos was performed by Web Logo program.
Expression patterns of LBD genes in Brassica rapa
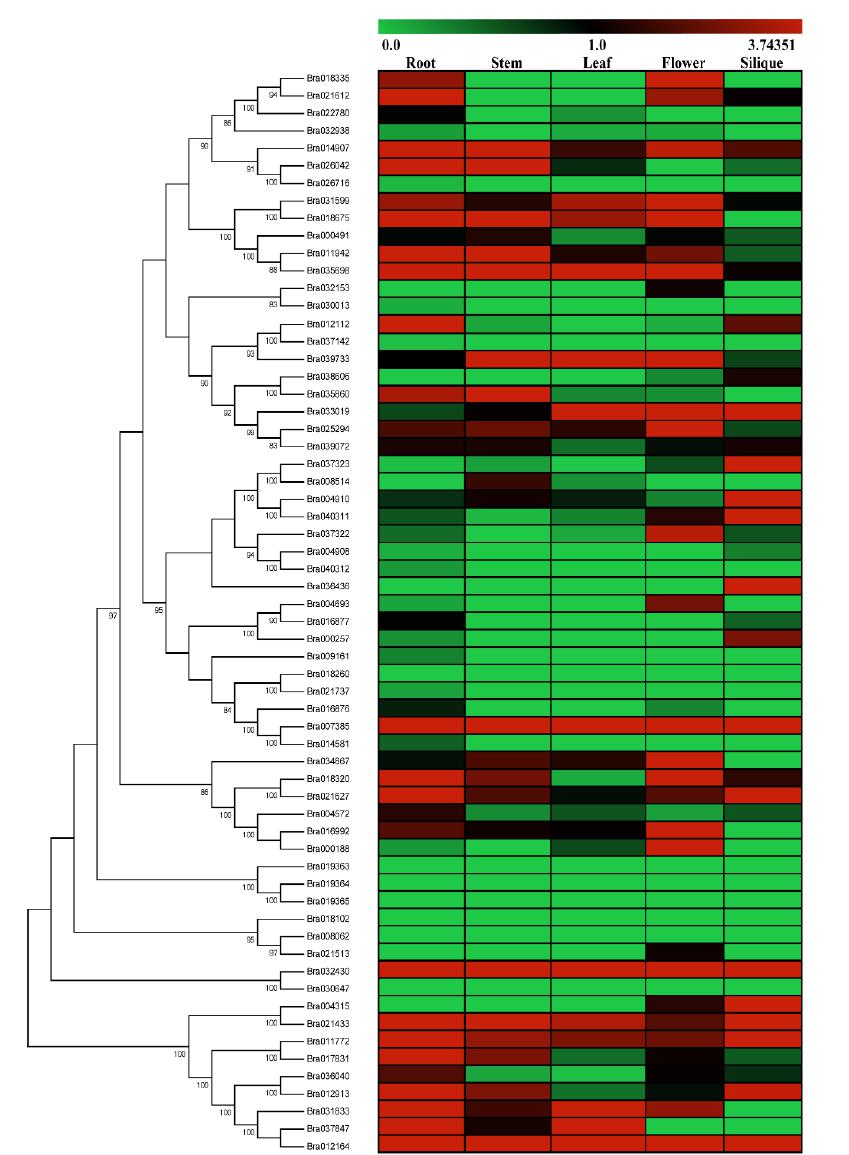
We analyzed expression levels of Brassica rapa LBD genes in five tissue using Illumina RNA-seq data (Fig. 5). The transcript levels (FPKM values) of all LBD genes were obtained from at least one tissue. Among 62 BrLBDs, 23 genes were expressed in all tissues and seven genes (Bra018262, Bra019363, Bra019364, Bra019365, Bra018102, Bra028062 and Bra030647) were not expressed in all tissues, while Bra017385 expressed at high level (FPKM>30) and Bra032430 expressed at high level (FPKM>10) in all tissues. In general, the expression level in each organ was as follows: root >flower >stem > silique > leave, and most genes of class II were expressed in each tissue and have a high expression level. Several genes exhibited tissue-specific expression, for example, Bra018335 and Bra004693 was expressed only in root and flower; Bra032153 and Bra021513 was expressed only in flower; Bra039606 and Bra004315 was detected only flower and silique; Bra018675, Bra034867 and Bra031833 were not detected silique. Interestingly, the FPKM value of Bra004315 exceeded 115 in the silique, demonstrating that it may be important in Brassica rapa. Furthermore, we also detected the expression of the duplicated genes, which had the similar gene structure and got together in the phylogenetic tree. However, the expressions of several duplicated genes were different. Detailed expression values and clusters of each LBD gene were analyzed using cluster analysis based on RNA-seq datasets (Fig. 5).
Expression profile cluster analysis of the Brassica rapa LBD family proteins. Expression values of each LBD gene identified in the study were downloaded from RNA-seq data, including roots, stems, leaves, flowers, and siliques.
DISCUSSION
Brassica rapa represents a nutritionally important vegetable whose genome has been fully sequenced. The complexity of the LBD gene family has been investigated in many plant species. However, there is little information on the biology and function of LBD proteins in Brassica rapa compared with other model plants, such as Arabidopsis and rice. The identification, classification, expression, and comparative analyses presented here provide a solid foundation for future studies of LBD protein regulatory functions during plant growth and development. This survey presents a comprehensive overview of the LBD gene family repertoire within the Brassica rapa genome.
Through genome-wide identification and subsequent comparative analysis, we identified 62 LBD genes in Brassica rapa. The number of LBD genes was significantly different from that of Arabidopsis (43), rice (35), maize (44) and tobacco (98), indicating that there is certain difference of LBD genes in different plants, but the number is not proportional to the size of the genome. However, the number of class II was 9, which was similar to that of Arabidopsis (7/43), rice (5/35), tomato (6/40), maize (7/44) and tobacco (13/98), indicating that LBD genes possesses high conservatism during species evolution44 Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel plant-specific gene family. Plant Physiol. 2002; 129(2): 747-761.,1010 Yang Y,Yu X,Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006; 39(1): 248-262.,1212 Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana and functional and molecular comparisons between AS2 and other family members. Plant J. 2009; 58(3): 525-537.,1313 Wang XF, Liu X, Su L, Sun YJ, Zhang S, Hao YJ, et al. Identification evolution and expression analysis of the LBD gene family in tomato. Scientia Agri Sin. 2013; 46(12): 2501-2513.,1818 Sun TT, Gong DP, Zhang L, Chen YQ, Zhao W, Xiang XH, et al. Genome-wide Sequence Identification and Expression Analysis of the LBD Gene Family in Nicotiana tabacum. J Plant Genet Resour. 2016; 17(2): 316-325..
Genetic structure analysis is useful to explore gene family evolution and gene repeat event information. Genetic structure analysis is useful to explore gene family evolution and gene repeat event information. The exon/intron structures of the LBD genes were shown in Fig. 1, a large number of LBD genes had one to three introns (95.16%, 59 of 62); 14 contained one intron; 39 contained two introns; 6 contained three introns; the other three genes (Bra032938, Bra021627 and Bra032430) had 5, 6 and 8 introns, respectively. The same type of genes have similar genetic structure, which is in accordance with Arabidopsis and rice and other angiosperms44 Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel plant-specific gene family. Plant Physiol. 2002; 129(2): 747-761.,1010 Yang Y,Yu X,Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006; 39(1): 248-262..
The chromosomal location showed that the LBD gene was unevenly distributed on each chromosome of Brassica rapa, indicating that the LBD gene may have been widely distributed in the genome of the common ancestor. A gene cluster of six genes is located on chromosome A09, and the three pairs of genes are closely linked on chromosome A05, which may be caused by tandem duplication. Therefore, it is suggested that all segmental duplication and transposition events have played roles in the evolution of LBD gene family in Brassica rapa.
In this study, the LBD gene family was analyzed in Brassica rapa and three other species. A total of 195 LBD genes were identified and analyzed in our study. Based on structural characteristics and on a comparison of the phylogenetic relationships among Brassica rapa, Arabidopsis, tomato, and maize, 62 BrLBD genes were fell into two major classes and four subclasses. Orthologs are genes in different genomes that have been create through speciation events, while paralogs are genes in the same genome created through gene duplication events4343 Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics. Annu Rev Genom Hum G. 2000; 1(1): 41-73.. With each LBD protein class, we identified 4 pairs of paralogous gene in Brassica rapa, and all paralogous genes appeared between chromosome, indicating that genome duplication likely occurred in the long evolutionary process. While 29 pairs of orthologous genes have been obtained through phylogenetic relationships, of which 22 pairs occurred between Brassica rapa and Arabidopsis, 3 pairs occurred between Brassica rapa and tomato, 2 pairs occurred between Arabidopsis and tomato, which showed closer genetic relationship between cabbage and Arabidopsis. As a result, orthologous genes in Brassica rapa, Arabidopsis, and tomato may possess similar function. In addition, Brassica rapa, Arabidopsis, tomato and maize were distributed in all groups of LBD family, which suggests that there are no monocot-specific lineages among LBDs, but the complexity of subclasses might be different in monocots and dicots. The isolation and identification of these LBD genes are likely to assist in clarifying the molecular genetic basis for Brassica rapa genetic improvement and also provide functional gene resources for constructing transgenic plants.
The gene chip research of model plants Arabidopsis is more comprehensive compared with cabbage, and a large number of its LBD genes have been validated. The phylogenetic tree between Arabidopsis and Brassica rapa revealed that the LBD gene of similar function may be clustered the same class or subclass, and LBD gene is highly conserved, which provide evidence for function prediction of Brassica rapa LBD genes. For instance, it shows that AtLBD37 (At5G67420), AtLBD38 (At3G49940) and AtLBD39 (At4G37540) are induced by nitrate and involved in anthocyanin synthesis and nitrate metabolism44 Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel plant-specific gene family. Plant Physiol. 2002; 129(2): 747-761., therefore these genes clustered together in Class II of Brassica rapa may have similar functions, and there are 2 pairs of orthologous genes (At5G67420 and Bra031833, At4G37540 and Bra011772). AtLBD3 (At1G16530), AtLBD4(At1G31320) in Arabidopsis are induced cytokinin4444 Naito T, Yamashino T, Kiba T, Koizumi N, Kojima M, Sakakibara H, Mizuno T. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES2 LIKE9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci Biotech Bioch. 2007; 71(5): 1269-1278., therefore their orthologous genes clustered together in Class Ic (Bra026716 and Bra014907) of Brassica rapa may participate in the cytokinin signaling process. The functional verification of LBD gene in Arabidopsis also demonstrated that class I is mainly involved in developmental regulation, class II plays a role in physiological and biochemical pathways during the growth and development of plants such as nitrogen formation and environmental response.
The expression pattern of a gene is often correlated with its functional, therefore we analyzed the expression of the Brassica rapa LBD genes using RNA-seq data from five tissues (root, stem, leaf, flower and silique). The results showed that 55 LBD genes of Brassica rapa all 62 LBD genes were expressed in at least one developmental stage. Among them, class I gene has high diverse tissue-specific expression, these genes play a unique role in the growth and development of Brassica rapa, such as the formation of lateral root and lateral lobe, flower development and fruit ripening process. While many class II genes have a higher expression level in almost every tissue, which is similar to that of Arabidopsis, rice, pepper and other plant LBD families 1010 Yang Y,Yu X,Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006; 39(1): 248-262.,1212 Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana and functional and molecular comparisons between AS2 and other family members. Plant J. 2009; 58(3): 525-537.,1717 Zheng ZF, Zhang YL, H C, Dai XZ, Liu F, Yuan ZH. Genome-wide Identification and Expressing Analysis of LBD Transcription Factors in Pepper. Acta Hortic Sin. 2016; 43(1): 683-694.. In addition,some genes are highly expressed in certain tissues such as Bra018335 and Bra004693 in root and flower organs, Bra032153 and Bra021513 in flower Bra039606 and Bra004315 in flower and silique. It is worth mentioning that Soly06g050430 is mainly expressed in the root1212 Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana and functional and molecular comparisons between AS2 and other family members. Plant J. 2009; 58(3): 525-537., and its orthologous gene Bra004572 was also expressed in root.
CONCLUSION
In this study, a total of 62 BrLBD genes were identified in the whole Brassica rapa genome. Based on their structural characteristics and phylogenetic comparisons, the 62 BrLBD genes were classified into two classes (class I and class II) and four subclasses (Ia, Ib, Ic, Id). The LBD genes were unevenly localized on all the Brassica rapa chromosomes. The expression of LBD genes based RNA-seq were different in five tissues, which showed highly diverse tissue-specific expression and functional diversity. The bioinformatics analysis results of this study will be useful for future gene cloning and functional studies, and for creating Brassica rapa cultivars with improved genetic traits.
REFERENCES
-
1Husbands A, Bell EM, Shuai B, Smith HMS, Springer S. LATERAL ORGAN BOUNDARIES: defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007; 35(19): 6663-6671.
-
2Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009; 151(3): 1377-1389.
-
3Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011; 16(1): 47-52.
-
4Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel plant-specific gene family. Plant Physiol. 2002; 129(2): 747-761.
-
5Landschaslz WH, Johnson PF, McKnight SL. The lucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1998; 240(4860): 1759-1765.
-
6Semiarti E, Ueno Y, Tsukay H, Iwakawa H, Machida C, Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001; 128(10): 1771-1783.
-
7Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, et al. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007; 51(2): 173-184.
-
8Sun SB, Song JP, Meng LS. ASYMMETRIC LEAVES2 gene a member of LOB/AS2 family of Arabidopsis thaliana causes an abaxializing leaves in transgenic cockscomb. Mol Biol Rep. 2012; 39(4): 4927-4935.
-
9Chalfun-Junior A, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family controls proximal-distal patterning in Arabidopsis petals. Plant Mol Biol. 2005; 57(4): 559-575.
-
10Yang Y,Yu X,Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006; 39(1): 248-262.
-
11Zhu QH,Guo AY,Gao G,Zhong YF,Xu M,Huang M,et al. DPTF:a database of poplar transcription factors. Bioinformatics. 2007; 23(10): 1307-1308.
-
12Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana and functional and molecular comparisons between AS2 and other family members. Plant J. 2009; 58(3): 525-537.
-
13Wang XF, Liu X, Su L, Sun YJ, Zhang S, Hao YJ, et al. Identification evolution and expression analysis of the LBD gene family in tomato. Scientia Agri Sin. 2013; 46(12): 2501-2513.
-
14Wang X, Zhang S, Su L, Liu X, Hao Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. Plos One. 2013; 8(2): e57044.
-
15Jia XT, Liu WX, Xie WG, Liu ZP, Liu ZM, Wang YR. Genome-wide analysis of the LBD transcription factor family in Medicago truncatula. Acta Bot Boreal-Occident Sin. 2014; 34(10): 2176-2187.
-
16Zhang YM,Zhang SZ,Zheng CC. Genome wide analysis of LATERAL ORGAN BOUNDARIES domain gene family in Zea mays. J Genet. 2014; 93(1): 79-91.
-
17Zheng ZF, Zhang YL, H C, Dai XZ, Liu F, Yuan ZH. Genome-wide Identification and Expressing Analysis of LBD Transcription Factors in Pepper. Acta Hortic Sin. 2016; 43(1): 683-694.
-
18Sun TT, Gong DP, Zhang L, Chen YQ, Zhao W, Xiang XH, et al. Genome-wide Sequence Identification and Expression Analysis of the LBD Gene Family in Nicotiana tabacum. J Plant Genet Resour. 2016; 17(2): 316-325.
-
19Gombos M, Zombori Z, Szécsényi M, Gyorgyey J. Characterization of the LBD gene family in Brachypodium: a phylogenetic and transcriptional study. Plant Cell Rep. 2017; 36(1): 61-79.
-
20Dolan L, Langdale JA. New insights into plant development in New England. Development. 2004; 131(21): 5215-5220.
-
21Fan M, Xu C, Xu K, Hu YX. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012; 22(7): 1169-1180.
-
22Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, et al. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell. 2014; 26(3): 1081-1093.
-
23Rubin G, Tohge T, Matsuda F, Saito K, Scheible W. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009; 21(11): 3567-3584.
-
24Albinsky D, Kusano M, Higuchi M, Hayashi N, Kobayashi M, Fukushima A, et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol Plant. 2010; 3(1): 125-142.
-
25Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007; 19(11): 3037-3057.
-
26Ikezaki M, Kojima M, Sakakibara H, Kojima S, Ueno Y, Machida C, et al. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 2010; 61(1): 70-82.
-
27Yordanov YS,Regan S,Busov V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell. 2010; 22(11): 3662-3677.
-
28Evans MMS. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell. 2007; 19(1): 46-62.
-
29Zhang J,Tang W,Huang Y,Niu X,Zhao Y,Han Y,et al. Down-regulation of a LBD-like gene,OsIG1,leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J Exp Bot. 2015; 66(1): 99-112.
-
30Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature genet. 2011; 43(10): 1035-1039.
-
31Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998; 23(10): 403-405.
-
32Wu X, Song C, Wang B, Cheng J. Hidden Markov model used in protein sequence analysis. J Biomed Eng. 2002; 19(1): 455-458.
-
33Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012; 40(D1): 302-305.
-
34Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004; 32(suppl 1): D138-D141.
-
35Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012; 40(W1): W597-W603.
-
36Goodstein DM, Shu S, Howson R, Hayes RD, Fazo J, Mitros T, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40(D1): 1178-1186.
-
37Liu RH, Meng JL. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas. 2003; 25(3): 317-321.
-
38Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Hereditas. 2007; 29(8): 1023-1026.
-
39Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011; 28(10): 2731-2739.
-
40Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31(13): 3497-3500.
-
41Tong C, Wang X, Yu J, Wu J, Li W, Huang J, et al. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics. 2013; 14(1): 689.
-
42Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free open-source system for microarray data management and analysis. Biotechniques. 2003; 34(2): 374-378.
-
43Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics. Annu Rev Genom Hum G. 2000; 1(1): 41-73.
-
44Naito T, Yamashino T, Kiba T, Koizumi N, Kojima M, Sakakibara H, Mizuno T. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES2 LIKE9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci Biotech Bioch. 2007; 71(5): 1269-1278.
Publication Dates
-
Publication in this collection
2018
History
-
Received
30 Jan 2018 -
Accepted
05 Oct 2018