Abstracts
This paper describes the construction and characterization of a Brazilian strain of bovine herpesvirus type 1.2a (BoHV-1.2a) with a deletion of the glycoprotein E (gE) gene. The deletion was introduced by co-transfection of a deletion fragment containing the 5´and 3´gE flanking regions and genomic DNA of wild type BoHV-1 into bovine cells. Isolation of gE deletion mutant was performed by immunoperoxidase staining with an anti-gE monoclonal antibody. Viral clones were plaque purified and further examined by restriction endonuclesase digestion and Southern blot hybridization. This gE deletion mutant will be evaluated as a vaccinal virus, in order to determine its potential use for a differential vaccine.
infectious bovine rhinotracheitis; BoHV-1; deletion mutant; differential vaccine
Este artigo descreve a construção e caracterização de uma amostra de um herpesvírus bovino tipo 1.2a (BoHV-1.2a) que apresenta uma deleção na região genômica que codifica a glicoproteína E (gE). A deleção gênica foi induzida através da co-transfecção de um fragmento de deleção, contendo as regiões 5´e 3´flanqueadoras da gE, com o DNA viral intacto de uma amostra viral isolada de um animal que apresentava doença respiratória. O isolamento do vírus gE negativo (gE-) foi realizado com auxílio da técnica de imunoperoxidase em que foi utilizado como anticorpo primário um anticorpo monoclonal anti-gE. O vírus gE- foi purificado e o DNA isolado desta amostra foi examinado através das técnicas de análise por enzimas de restrição e "Southern blot". Esta amostra gE- será avaliada como candidata para compor uma vacina diferencial contra a rinotraqueíte infecciosa dos bovinos.
rinotraqueíte infecciosa; BoHV-1; vírus deletado; vacina diferencial
Construction and characterization of a glycoprotein E deletion mutant of bovine herpesvirus type 1.2 strain isolated in Brazil
Construção e caracterização de uma amostra de BoHV-1.2 isolada no Brasil com uma deleção no gene da glicoproteína E
Ana C. FrancoI,II; Frans A.M. RijsewijkIII; Eduardo F. FloresIV; Rudi WeiblenIV; Paulo M. RoeheI,V
ICentro de Pesquisas Veterinárias Desidério Finamor, Eldorado do Sul, RS, Brasil
IIUniversidade Luterana do Brasil, Canoas, RS, Brasil
IIIInstitute for Animal Science and Health, Division of Infectious Diseases and Food Chain Quality, (ID-Lelystad), Lelystad, The Netherlands
IVCentro de Ciências Rurais, Universidade Federal de Santa Maria, Santa Maria, RS, Brasil
VInstituto de Ciências Básicas da Saúde, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brasil
Correspondence to Correspondence Instituto de Ciências Básicas da Saúde Universidade Federal do Rio Grande do Sul Caixa Postal 2076. 90001-970, Porto Alegre, RS, Brasil. E-mail: proehe@orion.ufrgs.br
ABSTRACT
This paper describes the construction and characterization of a Brazilian strain of bovine herpesvirus type 1.2a (BoHV-1.2a) with a deletion of the glycoprotein E (gE) gene. The deletion was introduced by co-transfection of a deletion fragment containing the 5´and 3´gE flanking regions and genomic DNA of wild type BoHV-1 into bovine cells. Isolation of gE deletion mutant was performed by immunoperoxidase staining with an anti-gE monoclonal antibody. Viral clones were plaque purified and further examined by restriction endonuclesase digestion and Southern blot hybridization. This gE deletion mutant will be evaluated as a vaccinal virus, in order to determine its potential use for a differential vaccine.
Key words: infectious bovine rhinotracheitis/infectious pustular vulvovaginitis, BoHV-1, deletion mutant, differential vaccine
RESUMO
Este artigo descreve a construção e caracterização de uma amostra de um herpesvírus bovino tipo 1.2a (BoHV-1.2a) que apresenta uma deleção na região genômica que codifica a glicoproteína E (gE). A deleção gênica foi induzida através da co-transfecção de um fragmento de deleção, contendo as regiões 5´e 3´flanqueadoras da gE, com o DNA viral intacto de uma amostra viral isolada de um animal que apresentava doença respiratória. O isolamento do vírus gE negativo (gE-) foi realizado com auxílio da técnica de imunoperoxidase em que foi utilizado como anticorpo primário um anticorpo monoclonal anti-gE. O vírus gE- foi purificado e o DNA isolado desta amostra foi examinado através das técnicas de análise por enzimas de restrição e "Southern blot". Esta amostra gE- será avaliada como candidata para compor uma vacina diferencial contra a rinotraqueíte infecciosa dos bovinos.
Palavras-chave: rinotraqueíte infecciosa/vulvovaginite infecciosa dos bovinos, BoHV-1, vírus deletado, vacina diferencial
INTRODUCTION
Bovine herpesvirus 1 (BoHV-1), a major pathogen of cattle, is the agent of infectious bovine rhinotracheitis/infectious pustular vulvovaginitis virus (IBR/IPV) and it is associated to a number of other clinical syndromes, including pustular balanopostitis, abortion, infertility and conjunctivitis (3). BoHV-1 strains can be subdivided into three distinct genotypes, BoHV-1.1, BoHV-1.2a and BoHV-1.2b (9). Genotypes 1.1 and 1.2a are usually more virulent and can be associated to abortions, while strains from genotype 1.2b are usually less virulent and have not been related to abortions (2,10). Although such genomic differences do reflect pathogenic and antigenic differences, to date there is only one recognized antigenic group (16).
The BoHV-1 genome consists of a double stranded DNA molecule of about 135 kb, composed of a unique long (UL) and a unique short regions (US), flanked by an internal (IR) and a terminal (TR) inverted repeat sequences (13). The viral genome encodes approximately 70 different proteins, of which eleven are glycoproteins. One of these glycoproteins, glycoprotein E (gE), is encoded by a gene located within the US region (11,17,18). Glycoprotein E is not essential for viral replication, but in vitro growth analysis of viral mutants show a decreased cell-to-cell spread and smaller plaque size for gE negative viruses (11,17). In addition, gE deletion in vivo has been related to a decreased virulence in calves (1,14,15).
Both spontaneous and artificially induced viral mutations have successfully been used in animal vaccine production. An european gE negative (gE-) BoHV-1.1 strain has been used as a differential vaccine for the control of BoHV-1 infections, allowing the differentiation between the immune responses of infected and vaccinated animals (1,14,15). Although BoHV-1 isolates are genetically stable, small antigenic variations may occur. Therefore, autochtonous isolates used for animal vaccine production may be more efficacious as vaccinal strains when compared to non-autochtonous strains. In this study we describe the deletion of the gE gene from a Brazilian isolate of BoHV-1.2a, a candidate strain for a differential vaccine.
MATERIALS AND METHODS
Virus strain and cells
The BoHV-1.2 strain SV265 (genotype 1.2a), isolated from an animal with respiratory disease during an outbreak of IBR in São Borja, Rio Grande do Sul, Brazil. The virus was multiplied in Madin Darby bovine kidney (MDBK) or in embryonic bovine trachea (Ebtr) cells. Cell cultures were kept in Eagle's minimal essential medium (EMEM) supplemented with 5% to 10% fetal calf serum, 0.05 % yeast lactalbumin hydrolysate, 2 mM glutamine and antibiotics (100 iu/ml penicillin, 100 µg/ml streptomycin and 20 iu/ml mycostatin).
Viral DNA extraction
Sub-confluent cells grown in 900 cm2 roller bottles were infected with virus at a multiplicity of infection of 0.1 to 1. Approximately 36 hours after infection, when cytopatic effect (CPE) was evident in 90-100% of the cells, the supernatant was removed, clarified at 5000 x g for 20 minutes and centrifuged at 100000 x g for two hours at 4°C. The viral pellet was resuspended in TE (Tris 10mM, EDTA 1mM, pH 7.4) and treated with sodium dodecil sulfate and proteinase K (final concentrations of 1% and 100 µg/µl, respectively) for one hour at 37°C. The viral DNA was extracted with equilibrated phenol, precipitated with ethanol, resuspended in TE pH 7.4 and stored at 4°C (12).
Polymerase chain reaction (PCR) and clonings
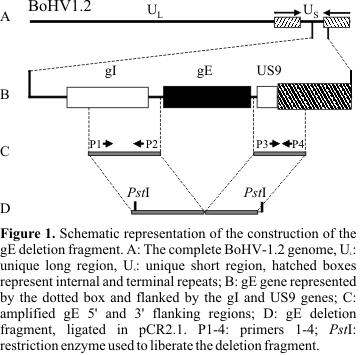
Amplification and cloning of the 5´ and 3´ gE flanking regions were performed to obtain the gE deletion fragment, according to the scheme shown in
Fig. 1. The primers used for amplification of the 5´region were P1: 5'-cgactgcttcgttatgctgc-3' and P2: 5'-GCGAGACCCATTTAACAACCC-3'. For the 3´ region, primers were P3: 5'- TGTGCCGTCTGACGGAAAGC-3' and P4: 5' -AATCCCCTCCTTCCCCTCC-3'. All primers sequences were obtained from the database (7). The predicted sizes of 5´ and 3´ gE flanking regions were 1100 base pairs (bp) and 900 bp, respectively, and the conditions for amplification were: 80°C for 30 seconds, 94°C for 1 minute, 60°C for 1 minute and 72°C for 1 minute. This cycling profile was repeated 15 times and followed by additional 23 cycles as described: 94°C for 1 minute, 60°C for 1 minute and 72°C for 1 minute. A final extension step at 72°C for 7 minutes was applied.All amplification reactions were performed using SV265 isolated DNA as template and both amplicons were examined by restriction enzyme analysis. Data obtained was compared with expected fragments based on previously reported BoHV-1 sequences (7). Amplicons were separately cloned in plasmid pCR 2.1 (TA Cloning Kit, Invitrogen), following the manufacturer's specifications. One of the clones containing the 5´gE flanking region was digested with restriction enzymes, isolated from an agarose gel and subsequently ligated in one of the 3´gE clones in order to produce the recombinant plasmid containing both gE flanking regions (gEdeletion fragment).
Construction and isolation of the gE- recombinant
Co-transfections were performed by the calcium phosphate method (4). Wild type viral DNA (2 µg) was co-transfected with 2 µg of the gEdeletion fragment in presence of carrier DNA (16 µg of salmon sperm DNA) in Ebtr cells.
Forty eight hours after co-transfection, plates were frozen and thawed to liberate cell-associated virions. Supernatants of co-transfection reactions were then used to infect 96 well plates with pre-formed Ebtr monolayers in order to isolate recombinant viruses. After visualization of cytopathic effect (CPE) on Ebtr cells, plates were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS; 8.5g NaCl, 1.55g Na2HPO4, 0.23g NaH2PO4, pH 7.2 per liter) and the immunoperoxidase monolayer assay (IPMA) was performed as described (6), using the anti-gE monoclonal antibody number 75 (5) as the primary antibody. Wells with unstained viral plaques were recorded, the supernatants collected and used to infect 96-well plates with Ebtr monolayers.
IPMA was performed until the complete isolation (that is, no stained viral plaques) of gE- virus from wild type virus. The gE- virus was then submitted to three additional cycles of plaque purification in 6-well plates under semi-solid medium (1% of agarose in EMEM).
Restriction enzyme analysis of wild type and gE- viral DNA
DNA from wild type and gE- virus was isolated as described above and restriction enzyme analysis was performed according to standard methods (12). Viral DNA was digested with HindIII and EcoRI, the fragments were separated on a 0.5 % agarose gel and stained with ethidium bromide following standard procedures (12).
Southern blot
Digested viral DNA was transferred to hybridization transfer membrane GeneScreen Plus (Biotechnology Systems) according to standard methods (12). The gE probe was the 2026 bp insert of plasmid p318 that harbors the complete gE open reading frame from the AluI site up to the HincII site. This fragment starts 52 nucleotides upstream the gE start codon and ends 130 nucleotides downstream the gE stop codon. The 2026 bp fragment was labeled with 32PdATP using a nick translation kit (Boehringer Manheim) according to the manufacturer's instructions.
Membrane hybridization was performed with 10 ng of the probe diluted in hybridization buffer, pH 7.2 (0.5 M sodium phosphate buffer, 1 mM EDTA, 7% SDS) at 65°C overnight. After two washing steps, the membrane was exposed to a storage phosphor screen (Molecular Dynamics) and the fluorogram was made using the fluorescence scanning system "storm" (Molecular Dynamics).
RESULTS
Construction of the gE- fragment
The PCR of the 5´gE flanking regions originated an amplicon of the expected size (1100 bp). This fragment, denominated 5'gE-, was cloned in pCR 2.1 and examined by restriction enzyme analysis for orientation. The amplicon corresponding to the 3´gE flanking region however, gave rise to a fragment of 750 bp, 150 bp shorter than the expected size. This fragment was examined by restriction enzyme analysis and results carefully compared with previous published nucleotide sequences of other BoHV-1 strains (7). After ensuring that, despite its shorter size, the fragment corresponded to the desired region, it was subcloned along with the 5'gE- region. Two clones in the correct orientation, containing the 5´-3´gE flanking regions, were selected and digested with restriction enzyme PstI to isolate the deletion fragment after gel electrophoresis. The deletion fragment contains, in total, about 1900 bp, corresponding to the 5´ and 3´gE flanking regions, separated by 80 nucleotides, which correspond to the plasmid polylinker. This 80 nucleotides fragment harbors two EcoRI sites, one immediately downstream the 5´gE flanking region and one immediately upstream the 3´gE region. According to this approach, the induced deletion starts 135 nucleotides upstream the gE start codon and ends 20 nucleotides downstream the gE stop codon.
Co-transfection and isolation of deleted virus
Co-transfection with viral DNA and the deletion fragment was performed in 6 well plates. About seven hundred viral plaques were obtained after co-transfection of Ebtr cells with wild type virus and the gE deletion fragment. Selection of gE- virus was made by IPMA after infection of co-transfection supernatants in preformed Ebtr monolayers in 96 well plates. Two wells showing unstained viral plaques were selected and their supernatants were used to infect new Ebtr monolayers. After a new IPMA, completely unstained CPE was detected (data not shown) and the gE negative virus was submitted to three rounds of plaque purification.
Restriction endonuclease analysis
The results of the restriction endonuclease analysis are shown in
Fig. 2, panel A. Digestion with HindIII showed the disappearance of the 7.2 kilobase (kb) fragment (HindIII L band) from the deleted viral DNA (panel A, lane 2). Due to the absence of the HindIII site in the gE gene, HindIII C and F bands also disappear in gE- viral DNA, giving rise to two new DNA fragments of about 22.2 kb (after fusion of HindIII C and L) and 19 kb (after fusion of bands F and L). Digestion of gE negative viral DNA with EcoRI showed that the 17.6 kb fragment (denominated EcoRI D fragment) disappears and gives rise to two additional bands with about 11.6 and 4.2 kb (not visible on gel) (panel A, lane 3). Such genomic alterations were compatible with the loss of the expected 1.8 kb fragment corresponding to the gE gene and the insertion of new EcoRI sites in the deletion fragment. No other major genomic alterations could be detected in wild type or deleted viral DNA.Southern Blot
Digested wild type and gE negative viral DNAs were blotted and probed with the whole gE open reading frame (Fig. 2, panel B). Wild type DNA showed specific hybridization with the expected fragments, which are HindIII C, F and L and EcoRI D bands (panel B, lanes 4 and 5, respectively). No specific hybridization was observed in gE negative viral DNA, except for a weak signal with the new HindIII 22.2 and 19 kb and with the new 4.2 EcoRI fragment. This weak reaction occurred due to the 130 nucleotides overlap of the gE probe downstream the gE stop codon.
DISCUSSION
The introduction of gene deletions, either naturally or by genetic engineering, has been used to provide potential candidate virus strains for vaccine development. Others have shown that gE- bovine herpesviruses may be suitable vaccine viruses in differential vaccines (1,14). Although BoHV isolates are regarded as stable, small antigenic variations may occur and it is possible that autochtonous isolates are more antigenically representative of viruses that are affecting a specific geografical region. Therefore, we developed an autochtonous gE- BoHV-1 so that, with the use of an anti-gE serological test, the immune response to the vaccinal virus can be differentiated from that induced by wild type virus, and yet retaining most of the antigenic characteristics common to local BoHV-1 isolates. Other gE- bovine herpesviruses have been described (1,11,15,18), however, this is the first report on the introduction of a whole gE deletion on a BoHV-1.2 strain. An option was made for a BoHV-1.2a in view of the fact that isolates of this subtype not only may cause respiratory disease but may also be involved in reproductive disorders, causing miscarriage (3,15). Consequently, this vaccinal candidate may induce protection against both respiratory and reproductive disease.
The induction of the gE deletion was investigated by restriction endonuclease and Southern blot analysis. The pattern of bands obtained by digesting the wild type viral DNA with HindIII and EcoRI was according to the restriction maps of the BoHV-1.2 K22 strain (8). Digestion of deleted viral DNA showed a new pattern of bands, with the disappearance of HindIII L band, which corresponded to the loss of a HindIII site inside the gE gene. Also due to the loss of this HindIII site, we observed the production of two new fragments after fusion of HindIII L with C and L with F bands. Genomic alterations in EcoRI digested DNA were according to insertion of additional EcoRI sites in the deletion fragment, giving rise to two new DNA fragments of about 11.6 and 4.2 kb. These results confirmed the induced deletion of the gE gene from the viral genome, as well as the absence of gE expression in infected cells. No additional genomic changes were found, indicating that no other major rearrangements were produced as a result of extra unintended recombination events.
Previous studies were performed to access the in vitro behavior of gE- BoHV-1.1 mutants (1,11). In order to determine the in vitro growth behavior of the gE- BoHV-1.2 and to make comparative analysis between BoHV-1.1 and BoHV-1.2, growth kinetics and viral plaque size of viruses will be determined. Also, the in vivo effect of infection with deleted virus will be determined by animal experimental infection, in order to access its potential as a vaccine candidate strain.
Submitted: March 26, 2001; Returned to authors for corrections: July 02, 2001; Approved: April 06, 2002.
- 1. Chowdhury, S.I.; Ross, C.S.D.; Lee, B.J.; Hall, V.; Chu, S. Construction and characterization of a glycoprotein E gene deleted bovine herpesvirus type 1 (BHV-1) Recombinant Virus. Am. J. Vet. Res., 60: 227-232, 1999.
- 2. Edwards, S.; Newman, R.H.; White, H. The virulence of british isolates of bovid herpesvirus 1 in relationship to viral genotype. Brit. Vet. J., 147: 216-231, 1991.
- 3. Gibbs, E.P.J. and Rweyemamu, M.M. Bovine herpesviruses. Part I. The Vet. Bull., 47: 317-343, 1977.
- 4. Graham, F.L. and van der Eb, A.J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virol., 52: 456-467, 1973.
- 5. Kaashoek, M.J.; Moerman, A.; Madic, J.; Weerdmester, K; Rijsewijk, F.A.M.; van Oirschot, J.T. An inactivated vaccine based on a glycoprotein E-negative strain of bovine herpesvirus 1 induces protective immunity and allows serological differentiation. Vaccine, 13: 342-346, 1995.
- 6. Kramps J.A.; Magdalena J.; Quak J.; Weerdmeester K.; Kaashoek M.J.; Maris-Veldhuis M.A.; Rijsewijk F.A.; Keil G.; van Oirschot J.T. A simple, specific, and highly sensitive blocking enzyme-linked immunosorbent assay for detection of antibodies to bovine herpesvirus 1. J. Clin. Microbiol., 32: 2175-81, 1994.
- 7. Leung-Tack, P.; Audonnet, J-C.; Riviere, M. The complete DNA sequence and the genetic organization of the short unique region (Us) of the bovine herpesvirus type 1 (ST strain). Virol., 409-421, 1994.
- 8. Mayfield, J.E.; Good, P.J.; van Oort, H.J.; Campbell, A.R.; Reed, D.E. Cloning and cleavage site mapping of DNA from bovine herpesvirus 1 (Cooper strain). J. Virol., 47: 259-264, 1983.
- 9. Metzler A.E.; Matile H.; Gassmann U.; Engels M.; Wyler R. European isolates of bovine herpesvirus 1: a comparison of restriction endonuclease sites, polypeptides, and reactivity with monoclonal antibodies. Arch. Virol., 85: 57-69, 1985.
- 10. Miller, J.M.; Whetstone, C.A.; Bello, L.J.; Lawrence, W.C. Determination of the ability of a thymidine kinase-negative deletion mutant of bovine herpesvirus-1 to cause abortion in cattle. Am. J. Vet. Res., 52: 1038-1043, 1991.
- 11. Rebordosa, X.; Piñol, J.; Pérez-Pons, J.A.; Lloberas, J.; Naval, J.; Querol, E. Mapping, cloning and sequencing of a glycoprotein-encoding gene from bovine herpesvirus type 1 homologous to the gE gene from HSV-1. Gene, 149: 203-209, 1994.
- 12. Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, New York, 1989.
- 13. Schwyzer M. and Ackermann M. Molecular virology of ruminant herpesviruses. Vet. Microb., 53: 17-29, 1996.
- 14. Strube, W.; Auer, S.; Block, W.; Heinen, E.; Kretzdorn, D.; Rodenbach, C.; Schmeer, N. A gE deleted infectious bovine rhinotracheitis marker vaccine for use in improved bovine herpesvirus 1 control programs. Vet Microbiol., 53: 181-9, 1996.
- 15. Van Engelenburg, F.A.C.; Kaashoek, M.J.; Rijsewijk, F.A.M.; van den Burg, L.; Moerman, A.; Gielkens, A.L.J.; van Oirschot, J.T. A glycoprotein E deletion mutant of bovine herpesvirus 1 is avirulent in calves. J. Gen. Virol., 75: 2311-2318, 1994.
- 16. Van Oirschot, J.T. Bovine herpesvirus 1 in semen of bulls and the risk of transmission: a brief review. The Vet. Quart., 17: 29-33, 1995.
- 17. Whitbeck J.C.; Knapp A.C.; Enquist L.W.; Lawrence W.C.; Bello L.J. Synthesis, processing, and oligomerization of bovine herpesvirus 1 gE and gI membrane proteins. J. Virol., 70:7878-84, 1996.
- 18. Yoshitake, N.; Xuan, X; Otsuka, H. Identification and characterization of bovine herpesvirus-1 glycoproteins E and I. J. Gen. Virol., 78: 1399-1403,1997.
Publication Dates
-
Publication in this collection
23 June 2003 -
Date of issue
Sept 2002
History
-
Accepted
06 Apr 2002 -
Received
26 Mar 2001 -
Reviewed
02 July 2001