Abstracts
Ribosomal operons are great tools for microbe community characterization and for microorganisms relationship study, particularly in the case of the acid lactic bacteria. The ribosomal operon of the probiotic strain Lactobacillus delbrueckii UFV H2b20 was partially characterized. A genomic library of this strain was constructed and the clones with partial ribosomal operon were sub-cloned using the shot-gun method for subsequent sequencing with the forward primer. The sequence analysis revealed that the 3' end of the rDNA 16S was following by the short spacer region 1 (16S-23S) and that the 3' end of the rDNA 23S was following by the short spacer region 2 (23S-5S), which preceded the rDNA 5S. In the flanking region of the rDNA 5S gene of this operon rrn, a region encoding six tRNAs was detected.
ribosomal operons; Lactobacillus; tRNA; acid lactic bacteria
Operons ribossomais têm sido instrumentos importantes na caracterização de comunidades microbianas e no estudo de relacionamentos entre microrganismos, principalmente em bactérias do ácido láctico. Operons ribossomais da linhagem probiótica, Lactobacillus delbrueckii UFV H2b20, foram parcialmente caracterizados. Um banco genômico da linhagem foi construído e os clones, contendo parte do operon ribossomal, foram subclonados pelo método de "shot gun", para em seguida serem seqüenciados com primer "forward". As seqüências indicaram a presença da extremidade 3' do rDNA 16S seguida da região espaçadora curta 1 (16S-23S) e a presença da extremidade 3' do rDNA 23S seguido da região espaçadora 2 (23S-5S), que por sua vez precedia o rDNA 5S. Adjacente ao gene rDNA 5S deste operon rrn uma região codificadora de 6 tRNAs foi detectada.
operons ribossomais; Lactobacillus; tRNA; bactérias do ácido lático
BIOTECHNOLOGY
Partial characterization of ribosomal operons of Lactobacillus delbrueckii UFV H2b20
Caracterização parcial de operons ribossomais de Lactobacillus delbrueckii UFV H2b20
Juliana Teixeira de MagalhãesI,* * Corresponding Author. Mailing address: Universidade Estadual de Santa Cruz UESC, Rodovia Ilhéus-Itabuna, Km 16, 45652-050, Ilhéus, BA, Brasil. Tel.: (+5573) 3680-5033, Fax: (+5573) 3680-5226. E-mail: julimaga@uesc.br ; Fernanda FlorestaII; Célia Alencar de MoraesII
ILaboratório de Genética e Biologia Molecular, Universidade Estadual de Santa Cruz, Ilhéus, BA, Brasil
IIDepartamento de Microbiologia, BIOAGRO, Universidade Federal de Viçosa, Viçosa, MG, Brasil
ABSTRACT
Ribosomal operons are great tools for microbe community characterization and for microorganisms relationship study, particularly in the case of the acid lactic bacteria. The ribosomal operon of the probiotic strain Lactobacillus delbrueckii UFV H2b20 was partially characterized. A genomic library of this strain was constructed and the clones with partial ribosomal operon were sub-cloned using the shot-gun method for subsequent sequencing with the forward primer. The sequence analysis revealed that the 3' end of the rDNA 16S was following by the short spacer region 1 (16S-23S) and that the 3' end of the rDNA 23S was following by the short spacer region 2 (23S-5S), which preceded the rDNA 5S. In the flanking region of the rDNA 5S gene of this operon rrn, a region encoding six tRNAs was detected.
Key words: ribosomal operons, Lactobacillus, tRNA, acid lactic bacteria
RESUMO
Operons ribossomais têm sido instrumentos importantes na caracterização de comunidades microbianas e no estudo de relacionamentos entre microrganismos, principalmente em bactérias do ácido láctico. Operons ribossomais da linhagem probiótica, Lactobacillus delbrueckii UFV H2b20, foram parcialmente caracterizados. Um banco genômico da linhagem foi construído e os clones, contendo parte do operon ribossomal, foram subclonados pelo método de "shot gun", para em seguida serem seqüenciados com primer "forward". As seqüências indicaram a presença da extremidade 3' do rDNA 16S seguida da região espaçadora curta 1 (16S-23S) e a presença da extremidade 3' do rDNA 23S seguido da região espaçadora 2 (23S-5S), que por sua vez precedia o rDNA 5S. Adjacente ao gene rDNA 5S deste operon rrn uma região codificadora de 6 tRNAs foi detectada.
Palavras-chave: operons ribossomais, Lactobacillus, tRNA, bactérias do ácido lático
INTRODUCTION
It has been established that phenotypic characteristics alone are not enough to allow a proper classification among the lactic acid bacteria. Thus, various reviews described the divergence of the results obtained using traditional phenotypic tests and the molecular taxonomy (10,11,15,25). The phylogenetic structure of this group is now well defined and is based on phenotypic division important instrument of group identification and classification as well as based on the data obtained from rRNA sequencing and DNA-DNA hybridization.
Ribosomal operons have acquired paramount relevance for the study of bacterial evolution and phylogeny. The 16S rRNA and 23S genes are the most widely used molecular chronometers for inferring microbial phylogeny and have been instrumental in developing a comprehensive view of the microbial systematic (9). The phylogenetic classification of prokaryotes with rDNA sequences is based on the assumption that the differences in sequences reflect the evolution of the organisms that they have been extracted from. For this reason, in the past few years, study based on the analysis of ribosomal sequences or genes encoding rRNA, were developed and used for to discriminate species of Lactobacillus or to identify different probiotic bacteria (18,24).
The sequences of multicopy rRNA genes are identical or nearly identical. The sequence of the different rrn operons existing in a given genome could vary up to 5% (4,16,23). The analysis of sequences found in the databank rrndb (Ribosomal RNA Operon Copy Number Database, http://rrndb.cme.msu.edu) revealed a variation of 1.23% between the operon sequences of E. coli and those of 14 others species (14).
The organization of the genes of the ribosomal operon is similar among the eubacteria. Basically, rrn operon contains the genes encoding the 16S, 23S and 5S ribosomal RNAs, which are organized as follows: 16S - spacer region 1 - 23S -spacer region 2 - 5S. The spacer regions are called "short" (S) when they do not encode any tRNAs, and called "long" (L) when a tRNA sequence is present (22). The presence of a tRNA encoding sequence within the spacer region 2 is not the nule in bacteria (5). Among the prokaryotes, the size, organization and number of rrn operons depends on the species. The highest copy number of rrn operons per genome (15 copy) was found in spore forming bacteria, like Clostridium paradoxum, while the lowest (one copy) was found in Mycoplasma pneumonie and Rickettsia prowazekii (13). In acid lactic bacteria, the number of operons varies from two in Lactobacillus brevis (14) to six in Lactococcus lactis (12,24) and Lactobacillus delbrueckii (17).
However, there is still limited knowledge of the organization of rrn operons in Lactobacillus delbrueckii. Because of its potential in the food industry as well as its clinical importance, the probiotic strain Lactobacillus delbrueckii UFV H2b20 has been studied in our laboratory. Preliminary analyses showed the existence of at least three copies of the rrn operon and polymorphism among the genes encoding the 16S rRNA. Intraspecific differences among industrial probiotic strains need to be established in order to monitor colonization in human and animal tests as well as their use in industrial products. We report here the partial characterization of a rrn operon of L. delbrueckii UFV H2b20 strain.
MATERIALS AND METHODS
Construction and screening of the genomic library of Lactobacillus delbrueckii
The genomic library of Lactobacillus delbrueckii UFV H2b20 was constructed in lambdaGEM®-11 phage (Promega - BamH I Arms, Cat. nº B1901) according to the manufacturer's instructions. The 16S rDNA of the strain was previously cloned in pGEM®-T Easy (Promega), labelled using the Primer-It Fluor Fluorescence Labeling Kit as described by the manufacturer (Statagene), and used as probe to screen the genomic library. The genomic library was transferred to Nylon membranes which were pre-hybridized for 2h at 65ºC in pre-hybridization buffer (SSC 5X, SDS 0.5%, Denhardt solution 5X, salmon sperm DNA 100 mg/mL) and hybridized 4h at 65ºC in the pre-hybridization buffer containing the rDNA 16S probe (15 ng/mL). Kodak X-OMAT K films were exposed to the membranes for 30 min at room temperature, and developed according to manufacturer's instructions. Insert of the positive clones were subject to phage DNA extraction and DNA was digested by SacI or XhoI according to the manufacturer's instructions (Promega), then analysed by Southern blot in the hybridisation conditions described above, using the 16S DNA probe. According to the restriction pattern and the Southern blot, five clones were selected and subcloned as 1.5 kb fragments in pBluescript by a shotgun method (Brazilian National Genome Project Consortium, 2003). Recombinant plasmids were used to transform the strain E coli (21).
Sequencing and sequence analysis
After transformation, about 200 colonies for each clone previously selected were pick-up and used for a subsequent sequencing. Plasmidic DNA was extracted and the inserts were sequenced with the primer forward M13 using the MegaBace 1000 DNA Analysis System (Molecular Dynamics & Life Science). Sequences were analysed using the Phred/Phrap software (www.phrap.org) and compared with the sequences available in the GenBank database (BLAST programs - www.ncbi.nlm.nih.gov). Contigs were defined by sequence overlapping as used for genome organization analysis (3).
RESULTS AND DISCUSSION
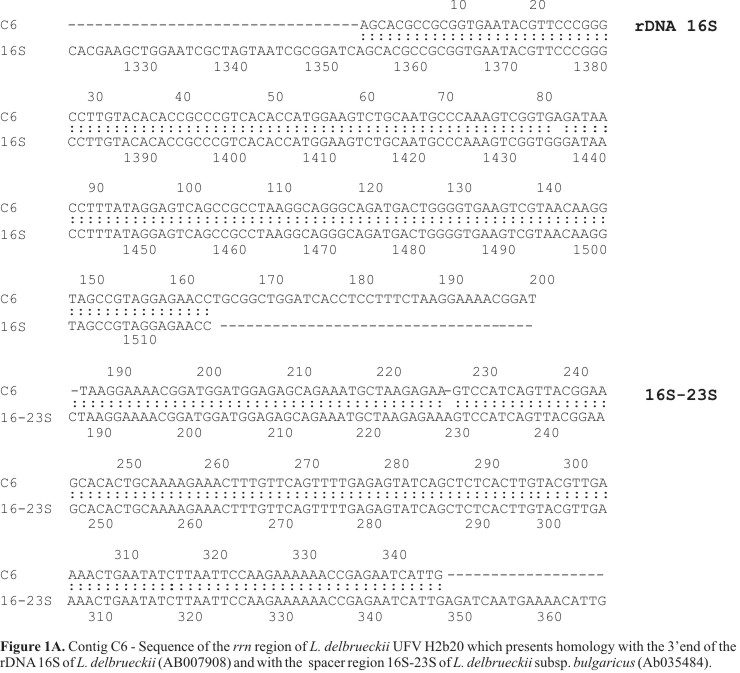
Based on the screening of a genomic library of L. delbrueckii using the rDNA 16S gene, 800 rrn operon sequences were obtained. These sequences were grouped in 50 different contigs with variable length (from 300 to 2000 pb). BLAST analysis of the contigs revealed a high homology with L. delbrueckii sequences already published (accession numbers: X15245, X68426, Z75476, AB035484, AF113602 and AB007908). We report here the analysis of two of these contigs, named C6 and C48. The contig C6 comprised 16S rDNA region, 16S-23S rDNA spacer, and the contig C48 comprised 23S rDNA, 23S-5S rDNA spacer, 5S rDNA region and tRNAs (Figs. 1A and B ).
The sequence analysis of L. delbrueckii UFV H2b20 rDNA showed that the rrn operon organization in this strain was similar to the one observed in the most eubacteria: the 3'end of the 16S rDNA preceded the spacer region 1; the 3'end of the rDNA 23S preceded the spacer region 2 that was followed by rDNA 5S. The Fig. 2 shows the organization of the rrn region of L. delbrueckii UFV H2b20.
It has been demonstrated that Lactobacillus acidophilus, Lactobacillus bulgaricus, Lactobacillus casei, Lactobacillus helveticus and Lactobacillus curvatus contained two main different types of rrn operon, named S and L. The rrnS operon was organized according to the following scheme: (5') 16S spacer region 1S 23S spacer region 2 5S (3'), while the rrnL operon was described as the following: (5') 16S spacer region 1L with (tRNAIle tRNAAla) 23S spacer region 2 5S (3') (18). In our study (Fig. 2), the spacer region 1 of L. delbrueckii UFV H2b20 was partially sequenced and was about 160 bp in length. It displayed 98.7% homology with the long spacer region 16S-23S of L. delbrueckii subsp. bulgaricus (accession number AB035484) but didn't show homology with the tRNAAla and tRNAIle encoding sequences observed in L. delbrueckii subsp. bulgaricus operons. Because the analysed sequence was partial 160 bp compared to the total length of the spacer region 1 of the Lactobacillus group which comprised between 197 and 220 bp we can not conclude if the spacer region 1 of L. delbrueckii UFV H2b20 contains or not any tRNA sequences. This was supported by the fact that, while most of the spacer regions 1 in the Lactobacillus group didn't contain tRNA sequences (8,17,18), they sometimes contain tRNAAla or tRNAIle or both. Sequence heterogeneity was found between the different rDNA encoded in the same genome (14) and the spacer regions were those which exhibited the highest degree of variation (2). The spacer region 23S-5S of L. delbrueckii UFV H2b20 was 71 bp in length and showed 95.7% homology with the corresponding region of L. delbrueckii (accession number X15245). The length of this region was in accordance with those of L. acidophilus (accession number Z75474; 69pb), L. bulgaricus (accession number Z75477; 71 pb) and L. helveticus (accession number Z75493; 71 pb). The obtained 5S rDNA of L. delbrueckii UFV H2b20 was 126 bp in length and displayed 98.6% of homology with the corresponding sequence of L. delbrueckii (accession number X15245). This length corresponds to the one observed for the L. lactis subsp. cremoris 117pb (1).
Sequences homologous to encoding elements associated to the rrn operons were also detected. We observed, at the 3' end of the rDNA 5S, 6 tRNA sequences which, in the following order, were homologous to the tRNAAsn, tRNAPro, tRNAGly, tRNAArg, tRNAVal and tRNAAsp of L. delbrueckii (accession number X15245) (Fig.1B ). Little information has been available about tRNA genes associated with rrn operon in Lactobacillus. However, in other species such as Bacillus subtilis, the 10 rrn operons were associated with groups of tRNAs: tRNAVal, tRNAThr, tRNALys, tRNALeu, tRNAGly, tRNAArg, tRNAPro, and tRNAAla were found located between the rrnJ and rrnW operons. In the 3' end of the rrnE operon, were found two tRNA genes, one for the methionine, the other for the aspartic acid (7, 20). In Staphylococcus aureus, the 3' ends of the main ribosomal operons were associated with tRNA encoding sequences (7). A tRNA for proline was also found beyond the rDNA 23S in the 3' end of an operon of Streptococcus mutans (19).
The special probiotic properties of L. delbrueckii UFV H2b20 have motivated the molecular characterization of this strain. Unique features of its rrn operons can be exploited to identify it among others of the same species, since rrn sequences are important tools for inter and intra-specific identification.
ACKNOWLEDGMENTS
Dr. J.T. Magalhães is supported by reseach grants from the FAPEMIG and the CNPq (Conselho Nacional de Desenvolvimento Científico). We are grateful to Dr. F. Micheli (CIRAD, Montpellier, France) for helpful discussion and advice on the manuscript. We also thank the team of the "Laboratório de Genética e Biologia Molecular" of the "Universidade Estadual de Santa Cruz" (UESC, Bahia, Brazil), and G.A.G. Pereira (UNICAMP, São Paulo, Brazil) for kindly providing assistance in the sequence analysis.
Submitted: April 05, 2005; Approved: June 28, 2005
- 1. Beresford, T.; Condon S. Physiological and genetic regulation of rRNA synthesis in Lactococcus.J. Gen. Microbiol., 139, 2009-2017, 1993.
- 2. Berthier, F.; Ehrlich, S.D. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett., 161, 97-106, 1998.
-
3Brazilian National Genome Project Consortium. The compete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci., USA, 100, 11660-11665, 2003.
- 4. Clayton, R.A.; Sutton G.; Hinkle, J.R.P.S.; Bult, C.; Fields C. Intraespecific variation in small - subunit rRNA sequences in genbank: Why single sequences may not adequately represent procaryotic taxa. Internat. J. System. Bacteriol., 45, 595-599, 1995.
- 5. Chen, H.; Lim, C.K.; Lee, Y.K.; Chan, Y.N. Comparative analysis of the genes encoding 23S-5S rRNA intergenic spacer regions of Lactobacillus casei-related strains. Interna. J. System. Evolution. Microbiol., 50, 471-478, 2000.
- 6. Green, C.J.; Vold, B.S. A cluster of nine tRNA genes between ribossomal gene operons in Bacillus subtilis.J. Bacteriol., 174, 3174-3151, 1992.
- 7. Green, C.J.; Vold B.S. Staphylococcus aureus has clustered tRNA genes. J. Bacteriol., 175, 5091-5096, 1993.
- 8. Gürtler, V.; Stanisich, V.A. New approaches to typing and identification of bacteria using the 16S-23S rRNA spacer region. Microbiology, 142, 3-16, 1996.
- 9. Gutell, R.R.; Larsen N.; Woese, C.R. Lessons from an involving rRNA structures from a comparative perspective. Microbiological Rev., 58, 10-26, 1994.
- 10. Hammes, W.P.; Weiss, N.; Holzapfell, W. The genera Lactobacillus and Carnobacteriu. In: Balows, A.; Truper, HG.; Dworkin, M.; Harder, W.; Schleifer, K. (eds). The prokaryotes, a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications 2nd ed. Volume II, Springer, New York, 1992, p. 1535-1573.
- 11. Kandler, O.; Weiss, N. Genus Lactobacillus Beijerinck 1901, 212. In: Sneath, P.H.H.; Mair, N.S.; Sharpe, M.E.; Holt, J.G. (eds). Bergey's Manual of Systematic Bacteriology. Williams e Wilkins, Baltimore, v. 2, 1986, p.12091234.
- 12. Klaenhammer, T.; Aaltermann, E.; Arigoni, F.; Bolotin, A.; Breidt, F.; Broadbent, J.; Cano, R.; Chaillou, S.; Deutscher, J.; Gasson, M.; Van de Guchte, M.; Guzzo, J.; Hartke, A.; Hawlkins, T.; Hols, P.; Hutkins, R.; Kleerebezem, M.; Kok, J.; Kuipers, O.; Lubbers, M.; Maguin, E.; Mckay, L.; Mills, D.; Nauta, A.; Overbeek, R.; Herman, P.; Pridmore, D.; Saier, M.; Van Sinderen, D.; Sorokin, A.; Steele, J.; O'Sullivan, D.; de Vos, W.; Weimer, B.; Zagorec, M.; Siezen R. Discovering lactic acid bacteria by genomics. Antonie van Leeuwenhoek, 82, 29-58, 2002.
- 13. Klappenbach, J.A.; Dunbar, J.M.; Schimdt, T. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environm. Microbiol., 66, 1328-1333, 2000.
- 14. Klappenbach, J.A.; Saxman, P.R.; Cole J.R.; Schimdt T. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res., 29, 181-184, 2001.
- 15. Klein, G.; Bonaperte, C.; Reuter G. Lactobacillus.In: Freney, J.; Reanud, F.; Hansen, W.; Bollet, C. (eds). Manuel de Bactériologie Clinique, vol II, 2nd ed. Elsevier, Amsterdam, 1994, p. 885-897.
- 16. Lan, R.; Reeves, P.R. Recombination between rRNA operons created most of the ribotype variation observed in the seventh pandemic clone of Vibrio colerae.Microbiology, 144, 1213-1221, 1998.
- 17. Moschetti, G.; Blaiotta, G.; Aponte, M.; Mauriello, G.; Villani, F.; Coppola, S. Genotyping of Lactobacillus delbrueckii subsp. bulgaricus and determination of the number and forms of rrn operons in L. delbrueckii and its subspecies. Res. Microbiol., 148, 501-510, 1997.
- 18. Nour, M. 16S-23S and 23S-5S intergenic spacer region of lactobacilli: nucleotide sequence, secondary structure and comparative analysis. Res. Microbiol., 149, 433-448, 1998.
- 19. Novak, J.; Shah, G.R.; Zhu, Y.; Caufield, P.W. Ribossomal RNA (rrn) operons in Streptococcus mutans and nucleotide sequence of tRNA (Pro) gene associated with rrnB. Oral Microbiol. Immunol., 12, 227-230, 1997.
- 20. Rudner, R.; Chevrestt, A.; Buchholz, S.R.; Studamire, B.; White, A.M.; Jarvis, E.D. Two tRNA gene clusters associated with rRNA operons rrnD and rrnE in Bacillus subtilis. J. Bacteriol., 175, 503-509, 1993.
- 21. Sambrook, J.; Fritsch, E.F.; Maniatis T. Molecular cloning. A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press, New York, 1989, p. 545.
- 22. Tannock, G.W. Identification of lactobacilli and bifidobacteria. In: Tannock, G.W. (ed). Probiotics, a critical review. Horizon Scientific Press, England, 1999, pp 45-56.
- 23. Tilsala-Timisjärvi, A.; Alatossava, T. Development of oligonucleotide of primers from the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int. J. Food Microbi., 35, 49-56.
- 24. Tulloch, D.L.; Finch, L.R.; Hillier, A.J.; Davidson, B.E. (1991) Physical map of the chromosome of Lactococcus lactis subsp. lactis DL11 and localization of six putative rRNA operons. J. Bacteriol., 173, 2768-2775, 1997.
- 25. Vandamme, P.; Pot, B.; Gillis, M.; de Vos, P.; Kersters, K.; Swings, J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiolog. Rev., 60, 407-438, 1996.
Publication Dates
-
Publication in this collection
16 Jan 2006 -
Date of issue
June 2005
History
-
Received
28 June 2005