Abstracts
Penicillium brevicompactum is a filamentous fungus that presents a potential for industrial use due its efficient pectinase production. A heterologous transformation system was developed for P. brevicompactum based on the complementation of a nitrate reductase mutant. Nitrate reductase mutants were obtained by resistance to chlorate in a rate of 23.24% when compared to other mutations that lead to the chlorate resistance. One mutant named 4457-18X was chosen for the transformation experiments with the pNH24 vector containing de Fusarium oxysporum nitrate reductase gene. A frequency of approximately 3 transformants/µg DNA was obtained using the circular vector pNH24. This frequency was multiplied about 10 fold using the linearized vector with the Xba I restriction enzyme. Southern analysis of the transformants showed a tendency of the linearized vector to diminish the number of integrations compared to the use of the circular vector. The integration was random and stable in the analyzed transformants. The establishment of a transformation system for P. brevicompactum is fundamental for genetic manipulation of this microorganism.
nitrate reductase; Penicillium brevicompactum; heterologous transformation
Penicillium brevicompactum é um fungo filamentoso que apresenta um potencial para a aplicação industrial devido a sua eficiente produção de enzimas do complexo pectinolítico. Neste trabalho foi desenvolvido um sistema de transformação heterólogo para P. brevicompactum baseado na complementação de um mutante nitrato redutase. Mutantes nitrato redutase foram obtidos pela resistência ao clorato de sódio em uma taxa de 23,24%. O mutante denominado 4457-18X foi escolhido para os experimentos de transformação com o vetor pNH24, que contém o gene da nitrato redutase de Fusarium oxysporum. Uma freqüência de cerca de 3 transformantes/mg de DNA foi obtida utilizando-se o vetor pNH24 na forma circular e um aumento de cerca de 10 vezes nessa freqüência foi alcançado com a utilização desse vetor linearizado com a enzima de restrição Xba I. A análise dos transformantes pela técnica de hibridização revelou uma tendência do vetor linearizado diminuir o número de integrações em relação ao vetor circular. A integração foi aleatória e estável nos transformantes analisados. O estabelecimento de um sistema de transformação para P. brevicompactum é essencial para a manipulação genética desse microrganismo.
nitrato redutase; Penicillium brevicompactum; transformação heteróloga
BASIC MICROBIOLOGY
Development of a transformation system for Penicillium brevicompactum based on the Fusarium oxysporum nitrate reductase gene
Desenvolvimento de um sistema de transformação para Penicillium brevicompactum baseado no gene da nitrato redutase de Fusarium oxysporum
Maurílio Antônio VaravalloI; Marisa Vieira de QueirozII,* * Corresponding Author. Mailing address: Departamento de Microbiologia, BIOAGRO, Universidade Federal de Viçosa, 36570-000, Viçosa, MG, Brasil. Tel.: (+5531) 3899-2553, Fax: (+5531) 3899-2573. E-mail: mvqueiro@mail.ufv.br ; Jorge Fernando PereiraII,III; Ronney Adriano RibeiroII; Marcos Antônio SoaresII; João Batista RibeiroII; Elza Fernandes de AraújoII
IFaculdade de Biomedicina, Centro Universitário das Faculdades Metropolitanas Unidas, São Paulo, SP, Brasil
IIDepartamento de Microbiologia, Instituto de Biotecnologia Aplicada à Agropecuária (BIOAGRO), Universidade Federal de Viçosa, Viçosa, MG, Brasil
IIIEmbrapa Trigo, Núcleo de Biotecnologia Aplicada a Careais de Inverno, Passo Fundo, RS, Brasil
ABSTRACT
Penicillium brevicompactum is a filamentous fungus that presents a potential for industrial use due its efficient pectinase production. A heterologous transformation system was developed for P. brevicompactum based on the complementation of a nitrate reductase mutant. Nitrate reductase mutants were obtained by resistance to chlorate in a rate of 23.24% when compared to other mutations that lead to the chlorate resistance. One mutant named 4457-18X was chosen for the transformation experiments with the pNH24 vector containing de Fusarium oxysporum nitrate reductase gene. A frequency of approximately 3 transformants/µg DNA was obtained using the circular vector pNH24. This frequency was multiplied about 10 fold using the linearized vector with the Xba I restriction enzyme. Southern analysis of the transformants showed a tendency of the linearized vector to diminish the number of integrations compared to the use of the circular vector. The integration was random and stable in the analyzed transformants. The establishment of a transformation system for P. brevicompactum is fundamental for genetic manipulation of this microorganism.
Key words: nitrate reductase, Penicillium brevicompactum, heterologous transformation
RESUMO
Penicillium brevicompactum é um fungo filamentoso que apresenta um potencial para a aplicação industrial devido a sua eficiente produção de enzimas do complexo pectinolítico. Neste trabalho foi desenvolvido um sistema de transformação heterólogo para P. brevicompactum baseado na complementação de um mutante nitrato redutase. Mutantes nitrato redutase foram obtidos pela resistência ao clorato de sódio em uma taxa de 23,24%. O mutante denominado 4457-18X foi escolhido para os experimentos de transformação com o vetor pNH24, que contém o gene da nitrato redutase de Fusarium oxysporum. Uma freqüência de cerca de 3 transformantes/mg de DNA foi obtida utilizando-se o vetor pNH24 na forma circular e um aumento de cerca de 10 vezes nessa freqüência foi alcançado com a utilização desse vetor linearizado com a enzima de restrição Xba I. A análise dos transformantes pela técnica de hibridização revelou uma tendência do vetor linearizado diminuir o número de integrações em relação ao vetor circular. A integração foi aleatória e estável nos transformantes analisados. O estabelecimento de um sistema de transformação para P. brevicompactum é essencial para a manipulação genética desse microrganismo.
Palavras-chave: nitrato redutase, Penicillium brevicompactum, transformação heteróloga
INTRODUCTION
Pectinolytic enzymes catalyze the degradation of the pectin present in the plant cell wall. Among these enzymes polygalacturonase preferentially hydrolyses pectic acids and pectin liase catalyses cleavage of the a-D-(1,4) glycosidic bounds of pectin by the beta-elimination mechanism. After analyzing the pectinase production of 10 species of the Penicillium genus, Pereira et al. (17) reported that Penicillium brevicompactum was the best pectin liase producer and presented a considerable polygalacturonase activity. Considering the important role that these enzymes have in some biotechnological process [for a review see Kashyap et al. (10)] this fungus became a promising organism for industrial use. For these applications, this species must be genetically improved to obtain an enhanced enzyme production and consequently a higher yield at an accessible cost.
Among the types of transformation systems used for filamentous fungi, the complementation of the nitrate reductase (niaD-) mutants offers a series of advantages. The spontaneous mutants can be easily obtained by positive selection by chlorate resistance and, since no mutagenic agents are used, the possibility of secondary mutations that would affect important genes is reduced. Besides, these mutants present a single desirable phenotype (inability to use nitrate as sole nitrogen source), being this phenotype not essential and not affecting growth or important metabolic pathways (28).
Heterologous and homologous transformation systems based on the complementation of the niaD- mutants have already been described for some filamentous fungi: Aspergillus oryzae (15,28), Fusarium oxysporum (5,13), Gibberella fujikuroi (23,27), Penicillium chrysogenum (7,35), and P. griseoroseum (16,19), among others. In the heterologous transformation systems, the transformation frequency was lesser than 20 transformants/µg DNA. The optimization of the heterologous transformation protocol for F. oxysporum reached a frequency of 100 to 200 transformants/µg DNA (11). One aspect that leads to an increase in the transformation frequency is the linearization of the vector. For several species, this linearization influenced the transformation frequency positively, as for Ustilago maydis with a 20-fold increase (34), A. niger and P. griseoroseum with a 2-fold increase (25,29). On the other hand, certain species, such as Coprinus cinereus, have not presented this effect (8).
One of the applications of a heterologous transformation system is the use as selection marker in cotransformation experiments to improve enzyme production, as for A. oryzae that presented up to 3.2-fold increases in polygalacturonase production when transformed with a P. janthinellum polygalacturonase gene, where the transformants had been selected by the complementation of a niaD- mutation (9), and to A. awamori, where nearly 90% of the transformants containing polygalacturonase encoding gene copies of A. niger presented a two to six-fold increase of this enzyme production (21). If this heterologous system shows random integrations, it can also be used for insertional mutagenesis experiments and future cloning of important genes in this fungus.
Thus, the goal of this work was to develop a heterologous transformation system for P. brevicompactum based on the complementation of the nitrate reductase gene that would allow the genetic manipulation of this species.
MATERIALS AND METHODS
Strain and media
The Penicillium brevicompactum strain was obtained from the Fundação Tropical de Tecnologia e Pesquisas André Tosello (Campinas/SP, Brazil) under the registration number CCT 4457. Minimum medium described by Pontecorvo et al. (18), complete medium according to Pontecorvo et al. (18) modified by Azevedo and Costa (2) and potato dextrose agar (PDA) with addition of 2.0 g peptone, 1.5 g hydrolyzed casein, 2.0 g yeast extract, and 1.0 mL vitamin solution (0.2 mg biotin, 10.0 mg p-aminobenzoic acid, 50.0 mg pyridoxine, 50.0 mg thiamine, 100.0 mg nicotinic acid, 100.0 mg riboflavin, distilled water for 100 mL) in 1000 mL of distilled water were used.
Transformation vector
Plasmid pNH24 was described by Diolez et al. (5) and has an insert of about 4.0 kb containing the Fusarium oxysporum nitrate reductase gene and unique sites for the enzymes Xba I and Eco RI.
Selection of chlorate resistant mutants
The selection of spontaneous mutants for the nitrate reductase gene (niaD-) was based on chlorate resistance as described by Unkles et al. (28). Approximately 1 x 106 conidia of the P. brevicompactum wild strain were plated in minimum medium containing sodium chlorate (470 mM) and glutamine (10 mM) as sole nitrogen source and incubated at 25ºC for 10 days. The chlorate resistant colonies were characterized on the basis of simple growth tests in minimum medium containing 10 mM of the following nitrogen sources: NaNO3, NaNO2, (NH3)2SO4, glutamate, and hypoxanthine (4). The colonies that presented growth in nitrite, hypoxanthine, glutamate and ammonium, but did not grow in NaNO3, were considered niaD- mutants.
Protoplast production and transformation
Protoplasts of P. brevicompactum were obtained as described by Varavallo (32). P. brevicompactum conidia were inoculated on Petri dishes with PDA medium, covered with cellophane paper, and incubated at 25ºC for 20 to 24 hours. The mycelium was removed and washed twice in osmotic stabilizer 0.8 M NaCl, prepared in 100 mM sodium and potassium phosphate buffer (pH 5.8). Approximately 300 mg of mycelium were incubated for 3 hours under agitation (80 rpm) at 30ºC in 5.0 mL of the osmotic stabilizer containing 15 mg.mL-1 of Glucanex enzyme (Novo Nordisk Ferment Ltd., Dittingen, Switzerland). The protoplasts were separated from the undigested mycelium by filtration, and washed twice in osmotic stabilizer STC (1.0 M Sorbitol, 50 mM CaCl2, 100 mM Tris-HCl) by centrifugation at 3000 g during 15 minutes at 4ºC. Finally, the pellet was resuspended in a certain STC volume for a final concentration of 1 x 107 protoplasts/0.1 mL.
The transformation was based on the method of Yelton et al. (33) and Balance and Turner (3). Approximately 2 x 107 protoplasts were mixed with 5.0 or 10.0 mg of plasmid pNH24 (circular or linearized with Xba I restriction enzyme) and 50 mL of 25% polyethylene glycol 6000 (PEG) and 50 mM CaCl2 in STC, homogenized and incubated on ice for 20 minutes. After this, 500 mL of the same PEG/CaCl2 solution were added and the mixture kept at room temperature during 20 minutes, then plated in minimum medium containing nitrate as sole nitrogen source and 0.8 M KCl as osmotic stabilizer. This was the best osmotic stabilizer to the P. brevicompactum protoplasts described by Varavallo (32). The negative control was realized with protoplasts that had been treated with PEG/CaCl2 solution, but without DNA. Controls of protoplasts regeneration were realized. All Petri dishes were incubated at 25ºC.
Mitotic stability
Mitotic stability of the niaD+ transformants was examined by growth on a non-selective medium (complete medium). Cultures were sequentially transferred to nonselective medium five times and cultured five days each time. Cultures were then transferred back to selective medium (minimum medium with nitrate as sole nitrogen source) to determine whether the selective marker had been lost.
Southern-blot analysis
For this analysis, the total DNA of three transformants obtained with the circular vector and three transformants obtained with the Xba I linearized vector was extracted according to the protocol of Specht et al. (26). Approximately, 3.0 µg of total DNA of each strain was used in the cleavage reactions with the Eco RI restriction enzyme. Electrophoresis and DNA transference to the nylon membrane (Stratagene) were carried out according to standard protocols (22). Plasmid pNH24 was used as probe. DNA probe labeling, hybridization and signal detection were performed with the "Dig High Prime Labeling and Detection Starter Kit II" (Roche) following the instructions of the manufacturer.
RESULTS AND DISCUSSION
Selection of nitrate reductase mutants
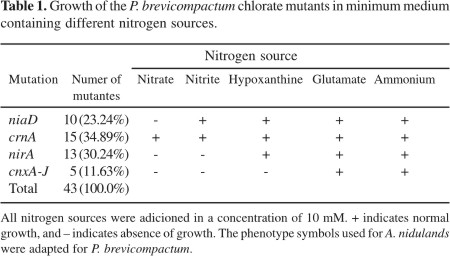
After 10 days of growth at 25ºC, 43 P. brevicompactum chlorate resistant mutants were selected (Table 1). Mutants with the chlorate resistance phenotype can be obtained by mutation in different genes (4), like the nitrate permease gene (crnA), the genes required for the biosynthesis of a molybdenum cofactor (cnxA-J), the specific and general regulators genes of the nitrate assimilation (nirA and areA) and in the proper nitrate reductase (niaD) gene. Independent of which gene is affected, mutation will prevent the cell from reducing chlorate to chlorite. These mutations can be easily differentiated on the basis of growth tests. Despite the phenotype of not growing in nitrate and nitrite could also stand in connection with the mutation of the nitrite reductase gene (niiA), there is no theoretical relation between the mutation in this gene and chlorate resistance.
Nitrate reductase mutants were obtained in a rate of 23.24% when compared to other mutations that lead to the chlorate resistance. Since the isolated mutants were spontaneous and because mutation is a random event, the number of isolated niaD- mutants for each species will predictably not follow a pattern, as reported for P. griseoroseum, where 8.33% of the chlorate resistants mutants were niaD- mutants (19), while in P. canescens the isolated niaD- mutants made up 49.0% (1), and 73.68% in P. chrysogenum (7).
After the isolation of niaD- mutants, the recipient strain for transformation procedures must be chosen. All the 10 niaD- mutants were tested for possible revertents and the reversion frequency was < 1 x 10-6 in all of them. The mutant designated 4457-18X was chosen for the transformation experiments since it presented the lowest residual growth in minimum medium containing nitrate as sole nitrogen source.
Transformation
Plasmid pNH24, which contains the F. oxysporum niaD gene, was used to transform the mutant 4457-18X. After two days at 25ºC, the transformants colonies were observed in selective medium containing nitrate as sole nitrogen source. This is a notably reduced time for the appearance of transformants. In comparison, the P. griseoroseum homologous niaD+ transformants were observed after five days (16), and for P. chrysogenum, 40% of the transformants appeared after seven days and the rest after the forty day (35). This growth speed seems to be an inherent characteristic of the P. brevicompactum and is of interest, since it diminishes the time required to obtain the desired transformants. All selected transformants transferred to a new selective medium were able to grow, without the presence of abortive transformants. Transformants were obtained in all tested treatments (Table 2).
The PEG treatment reduced the protoplast regeneration frequency by 9%, a similar rate reported for Metarhizium flavoviride with a 10% reduction of the regeneration rate (6) and for the CG423 strain of M. flavoviride with a 10 to 25% reduced protoplast viability (30).
The transformation frequency with the circular vector was around 3 transformants/mg DNA. This frequency is superior to the one obtained for G. fujikuroi of 1 to 2 transformants/µg DNA, using the A. niger niaD gene (23). However the frequency obtained in this study is lower than the one reported for P. griseoroseum of 13 transformants/µg DNA (25) or that reported by Ribeiro (20) of 19 transformants/µg DNA for P. expansum and reported by Levis et al. (12) of 10 transformants/µg DNA for Botrytis cinerea, being all of these also based on the complementation of a niaD- mutation with the pNH24 vector. Nevertheless, Langin et al. (11) presented a transformation frequency of 100 to 200 transformants/µg DNA for a heterologous system for F. oxysporum, by simply enhancing some aspects of the transformation protocol.
A number of studies have reported increased transformation frequency when using the vector in linear form. The integration of these linear vectors must occur by a non-homologous recombination mechanism, in which microhomologous regions of the simple strand ends of the vector would act in the integration. Enzymes like the topoisomerase I might be involved in this recombination type in S. cerevisae (36).
For P. brevicompactum the use of the linearized pNH24 vector with the Xba I restriction enzyme increased around 10 times the transformation frequency when using the concentration of 10 mg DNA. In U. maydis, the linearization of the vector pHL1 increased the transformation frequency by about 20 times (34), and studies into other fungi, such as A. bisporus,M. grisea, and S. cerevisae have reported the same observation (14,24,31). Whitehead et al. (35) showed that the use of the linearized vector pSTA10, which carries the A. niger niaD gene, increased the transformation frequency from 6.2 to 13.3 transformants/µg DNA. In the homologous transformation system of A. niger, the linearization of the vector increased the transformation frequency from 64 to 455 transformants/µg DNA (29).
Transformant analysis
After the monosporic purification, an analysis of the mitotic stability of the transformants was carried out. All analyzed transformants preserved their ability to use nitrate as sole nitrogen source after five transfers in complete medium. This indicates that the vector pNH24 integration was stable in the transformants. In heterologous transformation systems, vector integration is not always stable. Malardier et al. (13) demonstrated that some F. oxysporum transformants were instable after integrating the A. nidulans niaD gene into the genome.
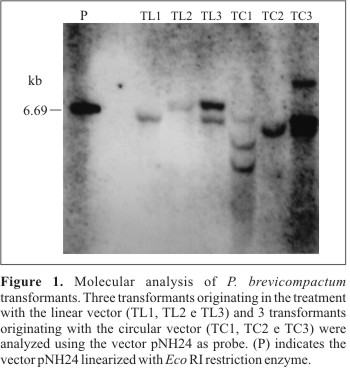
Hybridization experiments were realized with three transformants originated by the treatment with the circular vector and three by the treatment with the Xba I linearized vector. The physical presence of the vector was observed in all these transformants, indicating its integration in the genome (Fig. 1).
An analysis of the number of vector insertions into the transformant genome showed that there is a tendency of the linearized vector to reduce the number of integrations. Nevertheless, an analysis with a higher number of transformants is necessary to comprove this tendency. One vector copy seems to have been integrated in the transformants TL1, and TL2, while at two to three copies were integrated in the transformants TC1, and TC3. Soares (25) reported that two transformants originated from the transformation of P. griseoroseum with the circular plasmid pNH24 showed the integration of only one copy. However, this seems more likely to be an exception, as suggested by Queiroz et al. (19).
Unique and random integrations are the main requirements for the insertional mutagenesis technique, since they facilitate the verification of the integration into the gene that is responsible for the mutant phenotype and the future cloning of the interrupted gene. Queiroz et al. (19) reported that the greatest part of the pNE24 vector integrations in the P. griseoroseum genome occurred in heterologous sites. This characteristic was used by Soares (25) to develop an insertional mutagenesis system for P. griseoroseum using the REMI (Restriction Enzyme-Mediated Integration) technique, obtaining morphologic and auxotrophic mutants.
In conclusion, based on the results of this study, vector pNH24 can be used in an insertional mutagenesis system for P. brevicompactum, since this vector integrates randomly into the fungus genome and has a low number of copies when linearized.
ACKNOWLEDGMENTS
The authors thank the Brazilian agencies CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais) for financial support.
Submitted: May 05, 2004; Returned to authors for corrections: August 19, 2004; Approved: June 20, 2005
- 1. Aleksenko, A.Y.; Makarova, N.A.; Nikolaev, I.V. Integrative and replicative transformation of Penicillium canescens with a heterologous nitrate-reductase gene. Curr. Genet., 28, 474-477, 1995.
- 2. Azevedo, J.L.; Costa, S.O.P.; Exercícios práticos de genética. São Paulo, EDUSP: Editora Nacional, 1973, 288p.
- 3. Ballance, D.J.; Turner, G.; Development of a high-frequency transforming vector for Aspergillus nidulans.Gen., 36, 321-331, 1985.
- 4. Cove, D.J. Genetic studies of nitrate assimilation in Aspergillus nidulans.Biol. Rev., 54, 291-327, 1979.
- 5. Diolez, A.; Langin, T.; Gerlinger, C.; Brygoo, Y.; Daboussi, M.J.; The nia gene of Fusarium oxysporum: isolation, sequence and development of a homologous transformation system. Gene., 131, 61-67, 1993.
- 6. Furlaneto, M.C.; Paião, F.G.; Pinto, F.G.S.; Fungaro, M.H.P. Transformation of the entomopathogenic fungus Metarhizium flavoride to high resistance to benomyl. Can. J. Microbiol., 45, 875-878, 1999.
- 7. Gouka, R.J.; Hartingsveldt, W.V.; Bovenberg, R.A.L.; Hondel, C.A.M.J.J.; Gorcom, R.F.M. Cloning of nitrate-nitrite reductase gene cluster of Penicillium chrysogenum and use of the niaD gene as a homologous selection marker. J. Biotechnol., 20, 189-200, 1991.
- 8. Granado, J.D.; Chaloupková, K.K.; Aebi, M.; Kües, U. Restriction enzyme mediated DNA integration in Coprinus cinereus.Mol. Gen. Genet., 256, 28-36, 1997.
- 9. Ishida, Y.; Kakibuchi, K.; Hirao, Y.; Izumori, K. Cloning and characterization of a polygalacturonase-enconding gene from Penicillium janthinellum.J. Ferment. Bioeng., 84, 257-260, 1997.
- 10. Kashyap, D.R.; Vohra, P.K.; Chopra, S.; Tewari, R. Applications of pectinases in the commercial sector: a review. Biores. Technol., 77, 215-227, 2001.
- 11. Langin, T.; Daboussi, M.J.; Gerlinger, C.; Brygoo, Y. Influence of biological parameters and gene-transfer technique on transformation of Fusarium oxysporum.Curr. Genet., 17, 313-319, 1990.
- 12. Levis, C.; Fortini, D.; Brygoo, T. Transformation of Botrytis cinerea with the nitrate reductase (niaD) shows a high frequency of homologous recombination. Curr. Genet., 17, 157-162, 1997.
- 13. Malardier, L.; Daboussi, M.J.; Julien, J.; Roussel, F.; Scazzocchio, C.; Brygoo, Y. Cloning of the nitrate reductase gene (niaD) of Aspergillus nidulans and its use for transformation of Fusarium oxysporum.Gene., 78, 147-156, 1989.
- 14. Manivasakam, P.; Schiestl, R.H. Nonhomologous end joining during restriction enzyme-mediated DNA integration in Saccharomyces cerevisae.Mol. Cell. Biol., 18, 1736-1745, 1998.
- 15. Mattern, I.E.; Unkles, S.; Kinghorn, J.R.; Pouwels, P.H.; Van den Hondel, C.A. M.J.J. Transformation of Aspergillus oryzae using the A. niger pyrG gene. Mol. Gen., Genet. 210, 460-461, 1987.
- 16. Pereira, J.F. Caracterização e estudo da regulação do gene que codifica nitrato redutase de Penicillium griseoroseum e sua utilização como marcador de seleção para transformação homóloga. Viçosa, 2001, 37p. (MSc Thesis. Departamento de Microbiologia. UFV).
- 17. Pereira, J.F.; Queiroz, M.V.; Gomes, E.A.; Muro-Abad, J.I.; Araújo, E.F. Molecular characterization and evaluation of pectinase and cellulase production of Penicillium spp. Biotechnol. Lett., 24, 831-838, 2002.
- 18. Pontecorvo, G.; Roper, J.A.; Hemmons, L.M.; MacDonald, K.D.; Bufton, A.W.J. The genetics of Aspergillus nidulans. Adv. Genet., 5, 141-238, 1953.
- 19. Queiroz, M.V.; Barros, A.O.; Barros, E.G.; Guimarães, W.V.; Araújo, E.F. Transformation of Penicillium griseoroseum nitrate reductase mutant with the nia gene from Fusarium oxysporum.Can. J. Microbiol., 44, 1-3, 1998.
- 20. Ribeiro, J.B. Isolamento e caracterização de genes que codificam poligalacturonases e transformação de Penicillium expansum Viçosa, 2001, 57p. (MSc Thesis. Departamento de Microbiologia. UFV).
- 21. Ruttkowski, E.; Labitzke, R.; Khana, N.Q.; Loffler, F.; Gottschalk, M.; Jany, K.D. Cloning and DNA sequence analysis of a polygalacturonase cDNA from Aspergillus niger RH5344. Biochim. Biophys. Acta., 1087, 104-106, 1990.
- 22. Sambrook, J.; Russel, D.W. Molecular Cloning. A laboratory manual. 2nd ed.,.Cold Spring Harbor, Cold Spring Harbor Laboratory, New York, 2001.
- 23. Sànchez-Fernàndez, R.; Unkles, S.E.; Campbell, E.I.; Macro, J.A.; Cerda-Olmedo, E.; Kinghorn, J.R. Transformation of the filamentous fungus Gibberella fujikuroi using the Aspergillus niger niaD gene enconding nitrate reductase. Mol Gen. Genet., 225, 231-233, 1991.
- 24. Shi, Z.; Christian, D.; Leung, H.; Enhanced transformation in Magnaphorte grisea by restriction enzyme mediated integration of plasmid DNA. Phytopathology, 85, 329-333, 1995.
- 25. Soares, M.A. Mutagênese insercional e aumento da freqüência de transformação em Penicillium griseoroseum por meio de REMI (Restriction Enzyme Mediated Integration). Viçosa, 2002, 62p. (MSc Thesis. Departamento de Microbiologia. UFV).
- 26. Speacht, C.A.; Dirusso, C.C.; Novotny, C.P.; Ullrich, R.C. A method for extracting high molecular weight deoxyribonucleic acid from fungi. Anal. Biochem., 119, 158-163, 1982.
- 27. Tudzynski, B.; Mende, K.; Weltring, K.M.; Kinghorn, J.R.; Unkles, S.E. The Gibberella fujikuroi niaD gene encoding nitrate reductase: isolation, sequence, homologous transformation and electrophoretic karyotype location. Microbiology, 142, 553-539, 1996.
- 28. Unkles, S.E.; Campbell, E.I.; Rtuiter-Jacobs, Y.M.J.T.; Brockhuijsen, M.; Macro, J.A.; Carrez, D.; Contreras, R.; Van den Hondel, C.A.M.J.J.; Kinghorn, J.R. The development of a homologous transformation system for Aspergillus oryzae based on the nitrate reductase assimilation pathway: a convenient and general selection system for filamentous fungal transformation. Mol. Gen. Genet., 218, 99-104, 1989.
- 29. Unkles, S.E.; Campbell, E.I.; Carrez, D.; Grieve, C.; Contreras, R.; Fiers, W.; Van den Hondel, C.A.M.J.J.; Kinghorn, J.R. Transformation of Aspergillus niger with the homologous nitrate reductase gene. Gene., 78, 157-166, 1989.
- 30. Valadares-Inglis, M.C.; Inglis, P.W. Transformation of the entomopathogenic fungus, Metarhizium flavoviride strain CG423 to benomyl resistance. FEMS Microbiol. Lett., 155, 199-202, 1997.
- 31. Van de Rhee, M.D.; Graça, P.A.M.; Huizing, H.J.; MooibroeK, H. Transformation of the cultivated mushroom, Agaricus bisporus, to hygromycin B resistance. Mol. Gen. Genet., 250, 252-258, 1996.
- 32. Varavallo, M.A. Recombinantes com maior produção de pectinases e transformação em Penicillium spp. Viçosa, 2002, 78p. (Ph.D. Thesis. Departamento de Microbiologia. Universidade Federal de Viçosa).
- 33. Yelton, M.M.; Hamer, J.E.; Timberlake, W.E. Transformation of Aspergillus niger by using a trpC plasmid. Proc. Natl. Acad. Sci. USA., 81, 1470-1474, 1984.
- 34. Wang, J.; Holden, D.W.; Leong, S.A. Gene transfer system for the phytopatogenic fungus Ustilago maydis.Proc. Natl. Acad. Sci. USA., 85, 865-869, 1988.
- 35. Whitehead, M.P.; Unkles, S.E.; Ramsden, M.; Campbell, E.I.; Gurr, S.J.; Spence, D.; Van Den Hondel, C.; Contreras, R.; Kinghorn, J.R. Transformation of a nitrate reductase deficient mutant of Penicillium chrysogenum with the corresponding Aspergillus niger and A. nidulans niaD genes. Mol. Gen. Genet., 216, 408-411, 1989.
- 36. Zhu, J.; Schiestl, R.H. Topoisomerase I involvement in illegitimate recombination in Saccharomyces cerevisae.Mol. Cell. Biol., 16, 1805-1812, 1996.
Publication Dates
-
Publication in this collection
16 Jan 2006 -
Date of issue
June 2005
History
-
Received
20 June 2005