Abstracts
The aim of this study was to compare different DNA-extraction methods and selective enrichment broths for their effectiveness to detect Salmonella Typhimurium in artificially inoculated swine feces samples (100 CFU/g) by polymerase chain reaction. After enrichment in Rappaport-Vassiliadis, selenite cystine or Müller-Kauffmann tetrathionate, aliquots were used for DNA extraction by three different methods: boiling-centrifugation, phenol-chloroform and salting-out. Aliquots of extracted DNA were then used as template in PCR. The selective enrichment broths had no effect on the efficiency of PCR when boiling-centrifugation and salting-out were used. On the other hand, phenol-chloroform was superior (P<0.05) when combined to Rappaport-Vassiliadis. Considering cost and efficiency parameters, we encourage the use of Müller-Kauffmann tetrathionate broth in combination with boiling-centrifugation DNA-extraction procedure.
diagnosis; DNA; feces; PCR; Salmonella Typhimurium
O objetivo do presente estudo foi comparar diferentes técnicas de extração de DNA, realizadas a partir de três diferentes caldos de enriquecimento seletivo, na sua eficiência em detectar Salmonella Typhimurium em amostras de fezes suínas artificialmente inoculadas (100 UFC/g), pela técnica de reação em cadeia da polimerase (PCR). Após enriquecimento em Rappaport-Vassiliadis, selenito-cistina e tetrationato Müller-Kauffmann, alíquotas destes caldos foram utilizadas para extração do DNA, empregando três métodos diferentes, (a) fervura-centrifugação, (b) fenol-clorofórmio e (c) precipitação por sal. A eficiência dos métodos de extração de DNA por fervura-centrifugação e precipitação por sal foi a mesma, independentemente do caldo de enriquecimento seletivo utilizado. O caldo Rappaport-Vassiliadis apresentou maior eficiência (P<0,05) quando foi empregada a extração de DNA pelo método fenol-clorofórmio. Considerados os parâmetros custo e eficiência, os resultados do estudo indicaram que a partir de amostras fecais suínas a utilização do caldo tetrationato Müller-Kauffmann combinado a técnica de extração do DNA por fervura-centrifugação devam representar a melhor opção, relativamente às demais técnicas testadas.
diagnóstico; DNA; fezes; PCR; Salmonella Typhimurium
VETERINARY MICROBIOLOGY
Comparison of DNA-extraction methods and Selective Enrichment broths on the detection of Salmonella Typhimurium in swine feces by polymerase chain reaction (PCR)
Comparação entre métodos de extração de DNA e caldos de enriquecimento seletivo na detecção de Salmonella Typhimurium em fezes de suínos pela reação em cadeia da polimerase (PCR)
Carla Roberta FreschiI; Luiz Fernando de Oliveira e Silva CarvalhoI* * Corresponding Author. Mailing address: Departamento de Clínica e Cirurgia Veterinária, FCAVJ, UNESP, via de acesso Prof. Paulo Donato Castellane s/n, km 5, 14884-900, Jaboticabal, SP, Brasil. Tel.: (+5516) 3209-2688; fax: (+5516) 3202-4275. E-mail: lfosc@fcav.unesp.br ; Celso José Bruno de OliveiraII
IDepartamento de Clínica e Cirurgia Veterinária, Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista, Jaboticabal, SP, Brasil
IIDepartamento de Patologia Animal, Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista, Jaboticabal, SP, Brasil
ABSTRACT
The aim of this study was to compare different DNA-extraction methods and selective enrichment broths for their effectiveness to detect Salmonella Typhimurium in artificially inoculated swine feces samples (100 CFU/g) by polymerase chain reaction. After enrichment in Rappaport-Vassiliadis, selenite cystine or Müller-Kauffmann tetrathionate, aliquots were used for DNA extraction by three different methods: boiling-centrifugation, phenol-chloroform and salting-out. Aliquots of extracted DNA were then used as template in PCR. The selective enrichment broths had no effect on the efficiency of PCR when boiling-centrifugation and salting-out were used. On the other hand, phenol-chloroform was superior (P<0.05) when combined to Rappaport-Vassiliadis. Considering cost and efficiency parameters, we encourage the use of Müller-Kauffmann tetrathionate broth in combination with boiling-centrifugation DNA-extraction procedure.
Key words: diagnosis, DNA, feces, PCR, Salmonella Typhimurium
RESUMO
O objetivo do presente estudo foi comparar diferentes técnicas de extração de DNA, realizadas a partir de três diferentes caldos de enriquecimento seletivo, na sua eficiência em detectar Salmonella Typhimurium em amostras de fezes suínas artificialmente inoculadas (100 UFC/g), pela técnica de reação em cadeia da polimerase (PCR). Após enriquecimento em Rappaport-Vassiliadis, selenito-cistina e tetrationato Müller-Kauffmann, alíquotas destes caldos foram utilizadas para extração do DNA, empregando três métodos diferentes, (a) fervura-centrifugação, (b) fenol-clorofórmio e (c) precipitação por sal. A eficiência dos métodos de extração de DNA por fervura-centrifugação e precipitação por sal foi a mesma, independentemente do caldo de enriquecimento seletivo utilizado. O caldo Rappaport-Vassiliadis apresentou maior eficiência (P<0,05) quando foi empregada a extração de DNA pelo método fenol-clorofórmio. Considerados os parâmetros custo e eficiência, os resultados do estudo indicaram que a partir de amostras fecais suínas a utilização do caldo tetrationato Müller-Kauffmann combinado a técnica de extração do DNA por fervura-centrifugação devam representar a melhor opção, relativamente às demais técnicas testadas.
Palavras-chave: diagnóstico, DNA, fezes, PCR, Salmonella Typhimurium.
INTRODUCTION
Standard methods currently used to detect Salmonella clinical samples are laborious and require up to 7 days to obtain results. The polymerase chain reaction (PCR) assay represents a major advance in diagnostic methods in terms of speed and sensitivity. However, sample preparation prior to PCR is necessary, especially for feces, which contain high amount of compounds that are inhibitory for PCR (19). In spite of same enrichment broths to Salmonella are inhibitory for PCR (18), combination between different selective enrichment broths and PCR methods has been tested in order to improve sensitivity and dilute PCR inhibitory substances (6,15,18). The decision about the best methodology to be used to detect Salmonella in feces is very difficult due to the large amount of data that have been published. The aim of this study was to compare three DNA extraction methods (boiling-centrifugation, salting-out, phenol-chloroform) in combination with three different selective enrichment broths (Rappaport-Vassiliadis, Müller-Kauffmann tetrathionate and selenite cystine) for their effectiveness to detect Salmonella Typhimurium in artificially inoculated porcine fecal samples by PCR.
MATERIALS AND METHODS
Bacteria
Salmonella enterica subspecies enterica serovar Typhimurium (accession number 1007/00) previously isolated from swine feces was obtained from the culture collection of Instituto Adolfo Lutz, São Paulo, Brazil.
Inoculation of feces samples
Feces samples were collected from 10 sows of a farm where no Salmonella had been isolated before (12), stored in sterile plastic bags and taken to the laboratory under refrigeration to be processed on the same day. Each sample was subdivided in two 10-gram sub-samples. A fresh Salmonella Typhimurium culture prepared in tryptone soya broth was serially diluted and the number of Salmonella cells in each dilution was determined using the drop-counting technique (10). An aliquot (1 mL) of the dilution containing approximately 103 CFU/mL of Salmonella was added to one 10-gram sub-sample to obtain a final concentration of 102 CFU/g. The second 10-gram sub-sample was inoculated with 1 mL of phosphate-buffered saline (pH 7.4) as negative control.
Examination of spiked feces samples
Feces samples (10g) were pre-enriched in 90 mL of Hajna-GN broth (GN) at 37ºC for 6 hours. After pre-enrichment, 0.1 mL aliquot were transferred to 9.9 mL of Rappaport-Vassiliadis (RV), which were incubated at 42ºC for 24 hours. One mL aliquots of pre-enrichment broths were transferred to 9.0 mL of Müller-Kauffmann tetrathionate (MK) and 9.0 mL of selenite-cystine broth (SC), which were incubated at 37ºC for 24 hours and at 37ºC for 48 hours, respectively. After incubation, enrichment cultures were sub-cultured onto xylose-lysine-tergitol 4 (XLT4) agar plates, which were incubated overnight at 37ºC. Presumptive Salmonella colonies were inoculated on triple-sugar-iron agar and lysine-iron agar slants. All strains presumptively identified as Salmonella enterica were confirmed by slide agglutination test using poli-O and poli-H antiserum, as described by Poppoff and Le Minor (14).
DNA-extraction
The DNA extractions of enrichment broths were performed by following methods:
Boiling-centrifugation (M1) as described by Soumet et al. (17). One mL aliquots of enrichment broths were centrifuged 13,000 x g, 3 minutes. The pellets were resuspended in 100 mL of sterile bi-distilled water, heated to 95ºC in a dry block for 10 min, cooled in ice and centrifuged at 13,000 x g for 3 min. These supernatants were used for PCR assay.
Salting-out (M2) was carried out using a commercial kit (Puregene®, Gentra Systems) according to the instructions provided by the manufacturer.
Phenol-chloroform (M3) as described by Fadl et al. (4). Aliquots (1 mL) were centrifuged (2,000 xg, 4 minutes) and the pellet was resuspended in 474 mL of TE (10 mM Tris-HCL pH 8, 1 mM Na2EDTA), 25 mL 10% SDS and 1.25 mL proteinase K (20 mg/mL). After incubation at 55ºC for 30 minutes, 500 mL of phenol-chloroform pH 8 (1:1) was added, mixed vigorously and the samples were centrifuged (10,000 x g, 4 minutes). The aqueous phase was transferred to a fresh microtube and the DNA was precipitated with 3M sodium acetate and ice-cold isopropanol for 30 minutes. Samples were centrifuged (16,000 x g, 10 minutes) and the pellet was washed with 80% ethanol. The final pellet was resuspended in 50 mL of TE and stored at 4ºC until PCR was performed.
Oligonucleotide primers
The oligonucleotide primers S18 and S19 (Table 1) were based on the DNA sequence of the ompC gene of Salmonella enterica (9), used to amplify a 159bp fragment. The PCR product corresponded to base positions 1076-1234 bp of S. Typhi ompC gene (GenBank Accession Number M31424).
DNA amplification
The PCR mixture contained 5 mL of DNA sample, 2.0 mM MgCl2, 50 mM of each nucleotide, 0.4 mM of each primer and 1 U of Taq DNA polymerase in a final volume of 25 mL. Amplifications were performed in a thermal cycler (Personalâ, Eppendorf, Germany). After a 3-min denaturation at 96ºC, reaction mixtures were submitted to 10 cycles of amplification at 95ºC for 30 s, 62C for 30 s and 72ºC for 15 s, followed by 25 cycles at 95ºC for 30 s, 62ºC for 30 s and 72ºC for 30 s, and a final extension at 72ºC for 3 min. Amplified PCR products (10 mL) were then analysed by standard gel electrophoresis using 1.5% agarose gel and 1XTBE buffer (0.089 mol l-1 Tris-borate, 0.089 mol l-1 boric acid and 0.002 mol l-1 EDTA, pH 8.0), with a 100 bp ladder (Gibco-BRL, Gaithersburg, MD,USA) as molecular weight marker. Following electrophoresis, gels were stained with ethidium bromide (10 mg/mL) and visualized under U.V. light (254 nm).
PCR sensitivity
The PCR sensitivity was determined by using suspensions of Salmonella Typhimurium prepared in tryptone soya broth incubated overnight at 37ºC. Ten-fold serial dilutions were prepared of each selective enrichment broth and the numbers of bacteria were determined on plate count agar after incubation at 37ºC for 24 h. The same DNA-extraction procedures described above were carried out for each enrichment broth.
Statistical analysis
Each selective enrichment broth combined to DNA extraction method was repeated two separate times. The Binomial Test (16) was used to analyse the proportion of Salmonella positive samples in each selective enrichment broth associated with each DNA extraction method. Differences were considered significant at P<0.05.
RESULTS
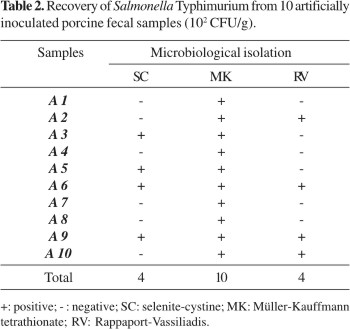
Significant differences (P<0.05) were found among the selective enrichment broths used for recovering Salmonella Typhimurium from feces samples. All samples were positive by culture using Müller-Kauffmann tetrathionate, whereas only 40% of the samples were positive using either RV or SC (Table 2).
No effects of selective enrichment broth on the efficiency of PCR were seen when the DNA-extraction methods M1 and M2 were used. However, RV was superior to SC and MK when DNA was extracted by M3 (Table 3). The samples non-Salmonella inoculated (negative controls) were all PCR negative and also negative by culture.
The PCR sensitivity was dependent of DNA-extraction methods and selective enrichment broths used. Higher sensitivity was obtained with M3, which detected 3.6x102 and 3.6x10 CFU/g of Salmonella Typhimurium in samples enriched in SC and RV, respectively. PCR sensitivity was markedly lower for all DNA-extraction methods when samples were enriched in MK (3.6x108 CFU/g).
DISCUSSION
Feces contain high amounts of compounds that are inhibitory for PCR. The effectiveness of PCR assay to detect Salmonella in feces depends not only on the DNA-extraction method and the selective enrichment broth used but also on the interaction between them. The extraction and purification of DNA can decrease the amount of inhibitory substances and the selective enrichment can increase the number of viable cells. In this study, three DNA-extraction methods in combination with three different Salmonella selective enrichment broths were tested. Müller-Kauffmann tetrathionate broth was significantly (P<0.05) more efficient than SC and RV in detecting Salmonella from culture and PCR when the DNA-extraction was performed by boiling-centrifugation or salting-out.
The RV broth has showed excellent results on the isolation of Salmonella in swine feces (5), become the only selective enrichment broth used at many laboratories that process this samples. Therefore, our results didn't corroborate with theses authors, since RV broth had a lower efficiency in the microbiological isolation. On the other hand, its efficiency was higher when associated to the PCR in any three DNA-extraction techniques. The sensitivity difference may be partially explained because the PCR detects both viable and nonviable Salmonella cells, whereas culture detects only viable organisms (2).
Soumet et al. (17) observed that SC was less inhibitory to PCR than MK and RV. However, the results this study indicate that either of three broths wasn't inhibitory to the PCR when the DNA-extraction method used was boiling-centrifugation or salting-out. Our results corroborate previous reports in which RV was non-inhibitory to PCR (5,11). In fact, similar results between selenite cystine and RV broths have been reported (8). Schrank et al. (15) compared MK broth with SC broth and concluded that PCR combined with MK broth was more sensitive. These data are in accordance with our findings.
Some authors award the inhibitory effects of RV and MK for the presence of MgCl2 and biliary salts, respectively, on it formulation (1,17). The commercial formulations of RV can differ greatly considering their magnesium chloride concentrations. Indeed, Peterz et al. (13) demonstrated that RV containing 40 g/L of MgCl2 was more inhibitory to the recovering of Salmonella than those containing 29 g/L. RV broth used in our laboratory (Oxoid, CM 866) contains 13.58 g/L of MgCl2, what can explain its low inhibitory effect.
PCR efficiency was lower (P<0.05) when either MK or SC was used for DNA template preparation by phenol-chloroform technique. Phenol-chloroform extraction eliminates biological contaminants in stool suspensions that may inhibit PCR, but this method appears to be unable to remove non-biological substances (18,19). Wilde et al. (19) reported that phenol-chloroform extraction failed to remove the inhibitors unless the specimens were further purified with chromatography medium CF11.
The sensitivity of PCR for pure Salmonella-cultures was high when the DNA-extraction method by phenol-chloroform was associated to samples enriched in the RV and SC broths. Müller-Kauffmann tetrathionate (MK) was highly inhibitory to PCR when it was performed on pure cultures, which corroborates previous findings (17). However, MK was not inhibitory when PCR was performed using artificially inoculated feces samples (Table 3), probably due to the higher number of salmonella organisms recovered by MK when compared to SC and RV (Table 2).
The efficiency of PCR associated to the boiling-centrifugation DNA-extraction method was very similar to those achieved by using the commercial kit (Table 2). Therefore, DNA samples prepared by boiling-centrifugation were more easily degraded and could not be stored for long periods at 4ºC. Commercial kits for DNA-extraction may be valuable tools when long-term storage of the extracted DNA is required (7). However, the speed and low cost of the boiling-centrifugation technique must be emphasized. In this study, we showed that RV, SC and MK can be successfully used as source of DNA for PCR when combined to boiling-centrifugation or salting-out. This information is valuable since more complex DNA extraction methods are expensive, time-consuming and laborious. Finally, SC and MK broths must be avoided when DNA-extraction is performed with phenol-chloroform.
ACKNOWLEDGEMENTS
Our research was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, proc. 01/05098-1).
Submitted: May 07, 2004; Returned to authors for corrections: March 21, 2005; Approved: November 22, 2005
- 1. Aabo, S.; Andersen, J. K.; Olsen, J. E. Research note: Detection of Salmonella in minced meat by the polymerase chain reaction method. Lett. Appl. Microbiol, 21, 180-182, 1995.
- 2. Cohen, N.D; Wallis, D.E; Neibergs, H.L.; McElroy, A.P.; McGruder, E.D.; DeLoach, J.R.; Corrier, D.E.; Hargis, B.M. Comparison of the polymerase chain reaction using genus-specific oligonucleotide primers and microbiologic culture for the detection of Salmonella in drag-swabs from poultry houses. Poult. Sci., 73, 1276-1281, 1994.
- 3. Eyigor, A.; Carli, K.T.; Unal, C.B. Implementation of real-time PCR to tetrathionate broth enrichment step of Salmonella detection in poultry. Lett. Appl. Microbiol., 34, 37-41, 2002.
- 4. Fadl, A.A.; Nguyen, A.V.; Khan, M.I. Analysis of Salmonella enteritidis isolates by arbitrarily primed PCR. J. Clin. Microbiol, 33, 987-989, 1995.
- 5. Feder, I.; Nietfeld, J.C.; Kelly, B.; Butine, M.D.; McNamara, P.; Chengappa, M.M. Evaluation of enrichment techniques for the isolation of Salmonella choleraesuis from swine feces. J. Microbiol. Methods, 33, 143-151, 1998.
- 6. Fluit, A.C.; Widjojoatmodjo, M.N.; Box, A.T.A.; Torensma, R.; Verhoef, J. Rapid detection of Salmonellae in poultry with the magnetic immuno-polymerase chain reaction. Appl. Environ. Microbiol., 59, 1342-1346, 1993.
- 7. Gibson, J.R.; McKee, R.A. PCR products generated from unpurified salmonella DNA are degraded by thermostable nuclease activity. Lett. Appl. Microbiol., 16, 59-61, 1993.
- 8. Kongmuang, U.; Luk, J.M.C.; Lindberg, A.A. Comparison of three stool-processing methods for detection of Salmonella serogroups B, C2, and D by PCR. J. Clin. Microbiol., 32, 3072-3074, 1994.
- 9. Kwang, J.; Littledike, E.T.; Keen, J.E. Use of the polymerase chain reaction for Salmonella detection. Lett. Appl. Microbiol., 22, 46-51, 1996.
- 10. Miles, A.A.; Misra, S.S. The estimation of the bactericide power of the blood. J. Hyg. (Cambrige), 38, 732-739, 1938.
- 11. Oliveira, S.D.; Santos, L.R.; Schuch, D.M.T.; Silva, A.B.; Salle, C.T.P.; Canal, C.W. Detection and identification of salmonellas from poultry-related samples by PCR. Vet. Microbiol., 87, 25-35, 2002a.
- 12. Oliveira, C.J.B.; Carvalho, L.F.O.S.; Domingues Júnior, F.J.; Menezes, C.C.P.; Fernandes, S.A.; Tavechio, A.T. Dunging gutters filled with fresh water had no effect on the prevalence of Salmonella enterica on Brazilian swine farms. Prev. Vet. Med., 55, 173-178, 2002b.
- 13. Peterz, M.; Wiberg, C.; Norberg, P. The effect of the incubation temperature and the magnesium chloride concentration on growth of Salmonella in homemade and commercially available dehydrated Rappaport-Vassiliadis broth. J. Appl. Bacteriol., 66, 523-528, 1989.
- 14. Poppoff, M.Y.; Le Minor, L. Antigenic formulas of the Salmonella serovars. WHO collaborating centre for reference and research on Salmonella, Paris, 1997.
- 15. Schrank, I.S.; Mores, M.A.Z.; Costa, J.L.A.; Frazzon, A.P.G.; Soncini R.; Schrank A.; Vainstein, M.H.; Silva, S.C. Influence of enrichment media and application of a PCR based method to detect Salmonella in poultry industry products and clinical samples. Vet. Microbiol., 82, 45-53, 2001.
- 16. Siegel, S. O caso de uma amostra. In: McGraw-Hill (ed). Nonparametric statistics for the behavioral sciences. New York, NY, 1975, pp. 39-66.
- 17. Soumet, C.; Ermel, G.; Fach, P.; Colin, P. Evaluation of different DNA extraction procedures for the detection of Salmonella from chicken products by polymerase chain reaction. Lett. Appl. Microbiol., 19, 294-298, 1994.
- 18. Stone, G.G.; Oberst, R.D.; Hays, M.P.; McVey, S.; Chengappa, M.M. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation-PCR procedure. J. Clin. Microbiol., 32, 1742-1749, 1994.
- 19. Wilde, J.; Eiden, J.; Yolken, R. Removal of inhibitory substances from faecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J. Clin. Microbiol., 28, 1300-1307, 1990.
Publication Dates
-
Publication in this collection
10 May 2006 -
Date of issue
Dec 2005
History
-
Received
07 May 2004 -
Reviewed
21 Mar 2005 -
Accepted
22 Nov 2005