Abstracts
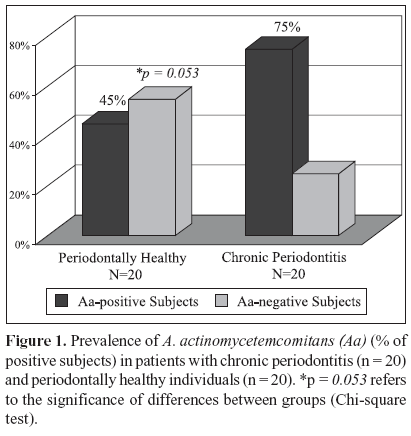
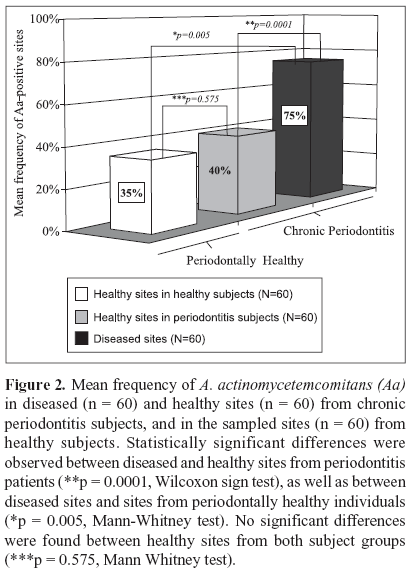
Actinobacillus actinomycetemcomitans is considered a major etiologic agent of aggressive periodontitis but this species has also been associated with other forms of periodontal disease. Further, highly leukotoxic strains are related to severity of disease. This investigation determined the prevalence of A. actinomycetemcomitans and the occurrence of the leukotoxin gene 530-bp deletion in Brazilian subjects with chronic periodontitis. Twenty periodontally healthy and 20 chronic periodontitis subjects were selected. Full-mouth clinical examination was carried out at 6 sites/tooth. Subgingival biofilm samples were collected from the 3 deepest sites and 3 healthy sites from periodontitis subjects, as well as from 3 sites of individuals with periodontal health. A. actinomycetemcomitans and the genetic deletion were determined by the polymerase chain reaction. Significant differences were sought by Mann-Whitney, Chi-square, and Wilcoxon sign tests. Periodontitis subjects presented a higher prevalence (75%) of A. actinomycetemcomitans than individuals with health (45%) (p = 0.053). A mean frequency of 57.5% of A. actinomycetemcomitans - positive sites was observed in the periodontitis group. Of those, 75% were diseased, whereas 40% were healthy sites (p = 0.0001). Healthy subjects showed a mean frequency of 35% of positive sites. In contrast, the genetic deletion was detected only in 4 diseased sites from 2 chronic periodontitis patients. A high prevalence of A. actinomycetemcomitans was observed in Brazilians with chronic periodontitis. However, the leukotoxin gene 530-bp deletion was rarely detected in the subgingival biofilm of these subjects.
chronic periodontitis; PCR; Actinobacillus actinomycetemcomitans; subgingival biofilm; leukotoxin
Actinobacillus actinomycetemcomitans tem sido associado com diferentes formas de doenças periodontais, mas tal espécie é considerada o principal agente etiológico da doença periodontal agressiva. Algumas cepas de A. actinomycetemcomitans apresentam uma deleção de 530 pb na região promotora do operon do gene da leucotoxina, produzindo assim maiores quantidades desta toxina. Tal fato pode ter um importante papel na patogênese das doenças periodontais. A proposta do presente estudo foi determinar a prevalência do A. actinomycetemcomitans em amostras de biofilme subgengival de indivíduos brasileiros com periodontite crônica e saúde periodontal; bem como avaliar a distribuição do tipo genético leucotóxico desta espécie nestes indivíduos. Vinte pacientes com periodontite crônica e 20 controles com saúde periodontal foram selecionados. O exame clínico periodontal foi realizado em 6 sítios/dente em todos os dentes. Amostras de biofilme subgengival foram coletadas de 3 sítios com a maior profundidade de bolsa e 3 sítios sem doença dos pacientes com periodontite, assim como de 3 sítios aleatórios dos controles. A detecção do A. actinomycetemcomitans e a ocorrência da deleção genética foram realizadas utilizando a técnica de PCR diretamente nas amostras de biofilme. Pacientes com periodontite crônica apresentaram uma alta prevalência de A. actinomycetemcomitans (75%) quando comparados aos indivíduos sem doença periodontal (45%), porém essa diferença não foi significativa (p = 0.053). Foi observada uma freqüência média de 57.5% de sítios com A. actinomycetemcomitans no grupo com periodontite. Destes sítios, 75% eram sítios com doença, enquanto 40% eram sítios saudáveis (p = 0.0001). Indivíduos sem doença periodontal apresentaram uma freqüência média de 35% de sítios com A. actinomycetemcomitans. A deleção de 530-pb foi encontrada somente em 4 sítios doentes de 2 pacientes com periodontite crônica. Uma alta prevalência de A. actinomycetemcomitans foi observada em pacientes Brasileiros com doença periodontal crônica. Entretanto, a deleção de 530 pb no gene da leucotoxina foi raramente detectada no biofilme subgengival desses indivíduos.
periodontite crônica; PCR; Actinobacillus actinomycetemcomitans; biofilme subgengival; leukotoxina
MEDICAL MICROBIOLOGY
Prevalence of leukotoxic genotypes of Actinobacillus actinomycetemcomitans in Brazilians with chronic periodontitis
Prevalência do genotipo leucotóxico de Actinobacillus actinomycetemcomitans em indivíduos brasileiros com periodontite crônica
Wilson Rosalem JuniorI,II; Arnaldo Feitosa Braga de AndradeI; Ana Paula Vieira ColomboIII,* * Corresponding Author. Mailing address: Rua General Dionísio, 60/604, Humaitá. 22271-050, Rio de Janeiro, RJ, Brasil. E-mail: anapaulacolombo@yahoo.com / apcolombo@micro.ufrj.br
IDepartamento de Microbiologia e Imunologia, Universidade Estadual do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
IIFaculdade de Odontologia, Universidade Estácio de Sá, Rio de Janeiro, RJ, Brasil
IIIDepartamento de Microbiologia Médica, Instituto de Microbiologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
ABSTRACT
Actinobacillus actinomycetemcomitans is considered a major etiologic agent of aggressive periodontitis but this species has also been associated with other forms of periodontal disease. Further, highly leukotoxic strains are related to severity of disease. This investigation determined the prevalence of A. actinomycetemcomitans and the occurrence of the leukotoxin gene 530-bp deletion in Brazilian subjects with chronic periodontitis. Twenty periodontally healthy and 20 chronic periodontitis subjects were selected. Full-mouth clinical examination was carried out at 6 sites/tooth. Subgingival biofilm samples were collected from the 3 deepest sites and 3 healthy sites from periodontitis subjects, as well as from 3 sites of individuals with periodontal health. A. actinomycetemcomitans and the genetic deletion were determined by the polymerase chain reaction. Significant differences were sought by Mann-Whitney, Chi-square, and Wilcoxon sign tests. Periodontitis subjects presented a higher prevalence (75%) of A. actinomycetemcomitans than individuals with health (45%) (p = 0.053). A mean frequency of 57.5% of A. actinomycetemcomitans - positive sites was observed in the periodontitis group. Of those, 75% were diseased, whereas 40% were healthy sites (p = 0.0001). Healthy subjects showed a mean frequency of 35% of positive sites. In contrast, the genetic deletion was detected only in 4 diseased sites from 2 chronic periodontitis patients. A high prevalence of A. actinomycetemcomitans was observed in Brazilians with chronic periodontitis. However, the leukotoxin gene 530-bp deletion was rarely detected in the subgingival biofilm of these subjects.
Key words: chronic periodontitis, PCR, Actinobacillus actinomycetemcomitans, subgingival biofilm, leukotoxin
RESUMO
Actinobacillus actinomycetemcomitans tem sido associado com diferentes formas de doenças periodontais, mas tal espécie é considerada o principal agente etiológico da doença periodontal agressiva. Algumas cepas de A. actinomycetemcomitans apresentam uma deleção de 530 pb na região promotora do operon do gene da leucotoxina, produzindo assim maiores quantidades desta toxina. Tal fato pode ter um importante papel na patogênese das doenças periodontais. A proposta do presente estudo foi determinar a prevalência do A. actinomycetemcomitans em amostras de biofilme subgengival de indivíduos brasileiros com periodontite crônica e saúde periodontal; bem como avaliar a distribuição do tipo genético leucotóxico desta espécie nestes indivíduos. Vinte pacientes com periodontite crônica e 20 controles com saúde periodontal foram selecionados. O exame clínico periodontal foi realizado em 6 sítios/dente em todos os dentes. Amostras de biofilme subgengival foram coletadas de 3 sítios com a maior profundidade de bolsa e 3 sítios sem doença dos pacientes com periodontite, assim como de 3 sítios aleatórios dos controles. A detecção do A. actinomycetemcomitans e a ocorrência da deleção genética foram realizadas utilizando a técnica de PCR diretamente nas amostras de biofilme. Pacientes com periodontite crônica apresentaram uma alta prevalência de A. actinomycetemcomitans (75%) quando comparados aos indivíduos sem doença periodontal (45%), porém essa diferença não foi significativa (p = 0.053). Foi observada uma freqüência média de 57.5% de sítios com A. actinomycetemcomitans no grupo com periodontite. Destes sítios, 75% eram sítios com doença, enquanto 40% eram sítios saudáveis (p = 0.0001). Indivíduos sem doença periodontal apresentaram uma freqüência média de 35% de sítios com A. actinomycetemcomitans. A deleção de 530-pb foi encontrada somente em 4 sítios doentes de 2 pacientes com periodontite crônica. Uma alta prevalência de A. actinomycetemcomitans foi observada em pacientes Brasileiros com doença periodontal crônica. Entretanto, a deleção de 530 pb no gene da leucotoxina foi raramente detectada no biofilme subgengival desses indivíduos.
Palavras-chave: periodontite crônica, PCR, Actinobacillus actinomycetemcomitans, biofilme subgengival, leukotoxina
INTRODUCTION
Over the years, a great diversity in the composition of the subgingival biofilm of various periodontal conditions has been shown (24). Many studies have attempted to clarify the relationship between a specific bacterium and/or a group of microorganisms and the pathogenesis of periodontitis. These data were originated in several parts of the world, including Europe, Africa, Asia, North and South America (5,8,9,15,23, 25,26,31,33). A general finding is that, even in different populations, the same periodontal species are found in sites or individuals with periodontitis. However, the prevalence and proportion of these species in the subgingival microbiota may vary (15,26). For instance, evidence has demonstrated a lower prevalence of Actinobacillus actinomycetemcomitans in the European and American than in the Asian population (1,4,9). Umeda et al. (36) also identified race or ethnicity as a potential risk factor for harboring oral A. actinomycetemcomitans. This species was detected more frequently in periodontal pockets of Hispanics and Asians-Americans when compared to Caucasians.
A. actinomycetemcomitans is a major periodontal pathogen that has been associated with aggressive forms of periodontitis. Nevertheless, this bacterial species can be found in 25% to 30% of chronic periodontitis lesions, even though the role of this microorganism in this type of periodontitis is not clear (30). A. actinomycetemcomitans elaborates a variety of toxins including a proteinaceous leukotoxin belonging to the repeats-in-toxin (RTX) family of bacterial cytolysins (20). This toxin kills human polymorphonuclear leukocytes and monocytes, but not epithelial and endothelial cells, fibroblasts, erythrocytes and platelets (32). The leukotoxin has membranolytic activity, producing pores in target cells with a functional diameter of approximately 0.96 nm (19). A. actinomycetemcomitans leukotoxin is expressed by an operon consisting of four genes designated ltxC, ltxA, ltxB and ltxD. The ltxA gene is essential for cytolytic activity since deletion of a repeat domain common to all RTX proteins in this region renders the leukotoxin inactive (12,21). Strains that demonstrate a 530-bp deletion in the promoter region of the leukotoxin gene operon exhibit enhanced leukotoxin production (17). Studies have shown an association between highly leukotoxic strains of A. actinomycetemcomitans and sites with destructive periodontal disease (17,18,34). Zambon et al. (41) detected the 530-bp deletion in A. actinomycetemcomitans isolates from 12 of 21 aggressive periodontitis patients but in none of the isolates from periodontally healthy and chronic periodontitis subjects. In addition to periodontal clinical status, different ethnic populations seem to show varying frequency of the 530-bp leukotoxin promoter deletion. Haubek et al. (17) found the genetic deletion in 37 of 84 A. actinomycetemcomitans strains from individuals of north and West Africa. In contrast, none of the isolates from subjects in north Europe presented the deletion (16).
Conventional methods used to identify periodontal pathogens in subgingival biofilm include culture techniques and biochemical testing (28). These techniques, however, are of limited specificity and sensitivity, quite expensive and time consuming (22). The polymerase chain reaction (PCR) method has been applied as a technique for the detection of A. actinomycetemcomitans in subgingival biofilm samples and saliva (35,36,39). Several studies have shown that this methodology appears to have a great potential in the microbiological diagnoses of periodontal diseases and detection of periodontal pathogens directly in subgingival biofilm samples (6,7,14,33,38).
In a previous study, we reported a very high prevalence of A. actinomycetemcomitans in Brazilians with chronic periodontitis, as well as periodontal health (5). These results raised the question whether the strains of A. actinomycetemcomitans present in the periodontitis patients were more virulent than the ones detected in the healthy individuals. Conceivably, diseased sites may harbor more leukotoxic strains than healthy sites. Thus, the purpose of the present study was to determine the prevalence of A. actinomycetemcomitans and the occurrence of the leukotoxin gene 530-bp deletion by PCR directly in subgingival biofilm samples of patients with chronic periodontitis from a Brazilian population.
MATERIALS AND METHODS
Subject population
Twenty periodontally healthy and 20 chronic periodontitis subjects were recruited from an ongoing study at the Dental School of the Estácio de Sá University (UNESA), Rio de Janeiro, Brazil. Subjects with chronic periodontal disease had at least 6 sites with probing pocket depth (PPD) and/or clinical attachment level (CAL) > 4 mm. Periodontally healthy individuals exhibited no sites with attachment loss or probing pocket depths > 4 mm. Subjects in this group, however, could exhibit minimal gingival inflammation. All patients were > 35 years of age and had at least 20 teeth. Exclusion criteria included pregnancy, use of antibiotics 6 months prior to entry into the study, or the presence of any systemic disease. In addition, patients with clinical symptoms and signs of aggressive periodontal disease, i.e. < 30 years of age and generalized interproximal attachment loss (PPD and CAL > 6 mm) affecting at least 3 permanent teeth other than first molars and incisors were excluded. The patients were informed about the nature of the study and a signed consent form was obtained from each individual. The study protocol was approved by the Review Committee for Human Subjects of UNESA (project no. 115).
Clinical measurements
Clinical measurements were performed at 6 sites per tooth at all teeth excluding third molars. They included PPD and CAL (mm) measured with a conventional North Carolina periodontal probe (Hu-Friedy, Chicago, IL), and presence or absence of supragingival biofilm accumulation (SB), bleeding on probing (BOP) and suppuration (SUP). All clinical measurements were performed by one calibrated examiner.
Sampling of subgingival biofilm
In the periodontitis group, individual subgingival biofilm samples were obtained from the 3 sites with the deepest PPD and from 3 healthy sites (presenting PPD < 3 mm and no signs of BOP), providing a total of 6 samples for each patient. In the periodontally healthy group, individual samples were obtained from 3 random healthy sites (PPD < 3 mm and no BOP). Prior to clinical measurements, supragingival biofilm was removed with sterile gauze from the selected sites. The sites were then isolated with cotton pellets and dried. A subgingival biofilm sample was taken from each site using a sterile curette and placed in separate tubes containing 200 µl of TE (Tris-HCl 10 mM, EDTA 1 mM, pH 7.6).
A. actinomycetemcomitans detection by PCR
Following subgingival biofilm sampling, bacterial DNA was extracted from each sample for detection of A. actinomycetemcomitans as described by Tan et al. (33). Briefly, bacterial cells were pelleted by centrifugation (2.500 x g for 2 min), the supernatant was discarded and the resulting pellet was resuspended in 50 µl of ultra pure water. This procedure was repeated 3 times. The final pellet was resuspended and bacterial cells were lysed by heating at 94ºC for 10 min, followed by cooling on ice for 5 min. Synthetic oligonucleotides (Invitrogen Brazil, São Paulo, SP, Brazil) targeting the leukotoxin gene ltxA of the ltx operon were used to detect A. actinomycetemcomitans by PCR. A. actinomycetemcomitans ATCC 29523 strain was used as positive control. The primers for ltxA used in this study were designed by Tonjum and Haas (35). The sequences of the forward and reverse primers were TT-15(5'-TCG CGA ATC AGC TCG CCG 3') and TT-16(5'-GCT TTG CAA GCT CCT CAC C-3'), respectively. Samples were amplified in a 50 µl reaction mixture, containing 10 ng of template DNA, 10 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2 (Biotools do Brasil Ltda., Rio de Janeiro, RJ, Brazil), 200 µM of each deoxynucleosidetriphosphate (Biotools), 0.5 µM solution of each primer, and 0.5 U of Taq polymerase (Biotools). PCR program was performed in a thermocycler (Primus 25/96, MWG - Biotech, Ebersberg, Germany), with a few modifications from the program described by Tan et al. (33). It included an initial denaturation cycle of 94ºC for 3 min, followed by 35 cycles of denaturation at 94ºC for 1 min, annealing at 60ºC for 1 min and extension at 72ºC for 1 min. The final extension was carried out at 72ºC for 10 min. The PCR products were analyzed by electrophoresis on a 1.5% agarose gel. The gel was treated with ethidium bromide (0.5 µg/mL) and visualized on an ultraviolet transilluminator. The presence of A. actinomycetemcomitans was determined by a distinct band of 285 bp in size. A pair of ubiquitous bacterial primers (5'-AGA GTT TGA TCC TGG CTC AG-3' and 5'-ACG GCT ACC TTG TTA CGA CTT- 3') (Biotools), designed by Willis et al. (40), which match almost all bacterial 16S RNA genes at the same position but not 18S RNA genes from eukaryotic cells, was used to indicate the presence of bacterial DNA in the clinical samples, particularly the ones that were A. actinomycetemcomitans- negative. For the reaction using universal 16S rDNA primers, PCR amplification was performed in a thermocycler (Primus 25/96, MWG - Biotech), and included a 25 µL reaction mixture containing 0.2 µM of forward and reverse universal primers, 2.5 µl of 10X PCR buffer (Biotools), 2 mM MgCl2, 1.25 U of Tth DNA Polymerase (Biotools), 200 µM of each deoxynucleosidetriphosphate (Biotools). The PCR temperature profile included an initial denaturation step at 97ºC for 1 min, followed by 30 cycles of a denaturation step at 97ºC for 45 s, a primer annealing step at 55ºC for 45 s, an extension step at 72ºC for 1 min and a final step of 72ºC for 4 min. The presence of bacterial DNA was determined by an amplicon of 1505 bp in size visualized on a 1.5 % agarose gel.
Determination of the leukotoxin gene promoter (ltx) type
In the A. actinomycetemcomitans-positive samples, PCR was carried out for determining the ltx promoter type. The set of primers A (5' - TCCATATTAAATCTCCTTGT-3') and B (5' - AACCTGATAACAGTATT-3') (Invitrogen Brazil) designed by Zambon et al. (41) were selected. Briefly, 100 ng of genomic DNA was mixed with 100 ng of each oligonucleotide primer, in the appropriate buffer. Samples were amplified in a 50 µL reaction mixture, containing 15 ng of template DNA, 10 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2 (Biotools), 200 µM of each deoxynucleotide triphosphate (Biotools), 50 pmol of each primer, and 1.5 U of Taq polymerase (Biotools). An initial denaturation cycle (95ºC for 5 min) was followed by 25 cycles of 30 s at 95ºC, 30 s at 55ºC, and 30 s at 72ºC. This was followed by a final cycle of 30 s at 95ºC, 30 s at 55ºC, and 9 min at 72ºC. These primers amplify a 492 bp product from highly toxic strains and a 1022 bp product from non leukotoxic strains. The amplicons were analyzed by electrophoresis on a 1.5% agarose gel. The gel was stained with ethidium bromide (0.5 µg/mL) and visualized on a ultraviolet transilluminator.
Statistical analysis
Full mouth clinical measurements were computed for each subject and then averaged across subjects within the groups. Differences on clinical parameters between the two groups were sought using the Mann-Whitney test. The frequency of detection of A. actinomycetemcomitans was determined for each group and significant differences between groups evaluated by Chi-square test. In addition, the mean frequency of A. actinomycetemcomitans was computed for each subject and averaged across subjects within the groups. Differences between groups were sought by Mann-Whitney test, whereas differences between healthy and diseased sites within the chronic periodontitis group were examined by the Wilcoxon sign test. Any difference of p < 0.05 was considered statistically significant.
RESULTS
The clinical features of the two groups are presented in Table 1. No significant differences between the groups were observed regarding age (p = 0.602) and gender (p = 0.337). In contrast, subjects with periodontitis presented significantly higher mean PPD and CAL, as well as mean number of missing teeth, % of sites with supragingival biofilm, bleeding on probing (p = 0.0001) and suppuration (p = 0.001; Mann-Whitney test) than periodontally healthy individuals. None of the selected subjects were smokers or past smokers for less than 5 years, although smoking was not an exclusion criterion.
The microbial data is depicted in Figs. 1 and 2. Of the 20 patients with chronic periodontitis, 15 (75%) were positive for A. actinomycetemcomitans; i.e. they presented at least 1 sampled site with this species, whereas in the periodontal health group 9 out 20 (45%) subjects were A. actinomycetemcomitans-positive. (Fig. 1; p = 0.053; Chi-square test). When the frequency of sites colonized by this species was determined for each subject and averaged within the groups, the mean prevalence of A. actinomycetemcomitans in the 60 samples from healthy individuals was 35% against 57.5% of the 120 sites from the chronic periodontitis group (p = 0.114; Mann - Whitney test; data not shown). Despite the fact that subjects and / or sites with periodontitis showed a greater prevalence of A. actinomycetemcomitans, no statistically significant differences between the groups were observed. However, when only the diseased sites (n = 60) from the chronic periodontitis subjects were considered, the mean frequency of A. actinomycetemcomitans was significantly higher in this group (75%) than in the healthy subjects (35%). (Fig. 2; p = 0.005, Mann-Whitney test). Similarly, a significantly greater prevalence of this species was observed in the diseased sites (75%) than in healthy sites (40%) from subjects with periodontitis (Fig. 2; p = 0.0001, Wilcoxon sign test). Although healthy sites from periodontitis subjects demonstrated a slightly higher mean frequency of A. actinomycetemcomitans (40 %) than healthy sites from individuals with periodontal health (35%), no significant difference between these groups was observed (Fig. 2; p = 0.575, Mann Whitney test). The analysis of the A. actinomycetemcomitans leukotoxin gene operon revealed that of all A. actinomycetemcomitans-positive samples (90 out of a total of 180 samples), only 4 samples from 2 diseased sites of 2 periodontitis subjects presented the 530 bp genetic deletion; i.e. the presence of strains with a highly leukotoxic profile. Interestingly, all the other sites from these 2 patients, including the diseased and healthy sites were positive for A. actinomycetemcomitans; however, the strains were not highly leukotoxic. All the biofilm samples that were negative for A. actinomycetemcomitans were positive for the 16S rRNA PCR amplification, confirming the presence of bacterial DNA in these samples.
DISCUSSION
The putative pathogen A. actinomycetemcomitans, which is mainly associated with aggressive forms of periodontitis, has also been frequently detected in sites and/or subjects with periodontal health and chronic periodontitis (30). In a previous study, Colombo et al. (5) found a high prevalence of A. actinomycetemcomitans in Brazilians, and speculated that different genotypes of this pathogen could predominate in chronic periodontitis or periodontal health. Thus, the present investigation determined the prevalence of this species, and the leukotoxic genotypes using the 530-bp leukotoxin operon deletion as a genetic marker, in chronic periodontitis and periodontally healthy subjects from a Brazilian population. The results here obtained showed a high frequency of A. actinomycetemcomitans in subjects and/or sites with chronic periodontitis, as well as periodontal health, confirming the previous data. Likewise, Avila-Campos and Velásquez-Meléndez (3) reported a prevalence of A. actinomycetemcomitans of 70% for healthy and 90% for chronic periodontitis Brazilians. Data from earlier investigations in Chinese, African and Hispanic populations are also in accord with these findings (2,8,29,33). In contrast, A. actinomycetemcomitans was detected in much lower frequency in Japanese, Turkish, Swedish and American subjects (10,11,15,37). Although different methodologies were employed in these investigations, they clearly demonstrated that the prevalence rates of this pathogen seem to vary among ethnic groups with various periodontal status. The widespread detection of this species in individuals with periodontal health and chronic periodontitis may suggest that this organism could be a member of the oral indigenous microbiota that occasionally contributes to the development of periodontitis. Nevertheless, the real role of this species in the etiopathogenesis of chronic periodontal disease is not fully understood.
Differences among these microbiological studies may also result from the use of distinct microbiological techniques. In the first study by our group, we analyzed the microbiota of Brazilians by whole genomic DNA probes (5). The major concern about this method was the high probability of false-positive results. Therefore, in the present study we employed the PCR technique due to the fact that it is rapid, highly sensitive and specific for the detection of periodontal pathogens (27). Indeed, a higher prevalence of A. actinomycetemcomitans was observed when the PCR method was employed in comparison to the use of whole genomic probes.
Although the presence of A. actinomycetemcomitans was demonstrated in 50% of all the periodontal sites evaluated (90/180 sites), only 4 samples from 2 diseased sites of 2 periodontitis subjects presented the 530-bp deletion. Thus, most of the strains found in those individuals with chronic periodontitis and/or periodontal health may be considered low leukotoxin producers. Similar results were reported by Cortelli and co-workers (6,7) in a Brazilian population with chronic periodontitis. Other studies have indicated that the deletion on the leukotoxin promoter region seem to be related to aggressive forms of periodontitis, particularly in subjects with close relationship to the African continent (13,17,41). Therefore, it is conceivable that the patients or sites in the current study harboring the leukotoxic clone are at higher risk to develop a more severe form of periodontal disease. Regarding the racial background, no reliable criteria to classify the ethnic groups were available due to the high miscegenation in this population. Nevertheless, no African-Brazilians were present in this sample.
In conclusion, a high prevalence of A. actinomycetemcomitans was observed in patients with chronic periodontitis from a particular Brazilian population. However, the leukotoxin gene 530-bp deletion was rarely detected in the subgingival biofilm of these subjects.
ACKNOWLEDGMENTS
This work was supported in part by Program of Research Support for Groups of Excellence (PRONEX), National Council for Scientific and Technological Development (CNPq), Foundation for Research Financial Support in the State of Rio de Janeiro (FAPERJ), Brazil.
Submitted: September 01, 2005; Returned to authors for corrections: March 09, 2006; Approved: July 18, 2006
- 1. Ali, R.W.; Velcescu, C.; Jivanescu, M.C.; Lofthus, B.; Skaug, N. Prevalence of 6 putative periodontal pathogens in subgingival plaque samples from Romanian adult periodontitis patients. J. Clin. Periodontol, 23, 133-139, 1996.
- 2. Ali, R.W.; Johannessen, A.C.; Dahlén, G.; Socransky, S.S.; Skaug, N. Comparison of the subgingival microbiota of periodontally healthy and diseased adults in Northern Cameroon. J. Clin. Periodontol, 24, 830-835, 1997.
- 3. Avila-Campos, M.J.; Velasquez-Melendez, G. Prevalence of putative periodontopathogens from periodontal patients and healthy subjects in São Paulo, Brazil. Rev. Inst. Med. Trop. São Paulo, 44, 1-5, 2002.
- 4. Choi, B.K.; Park, S.H.; Yoo, Y.J. Detection of major putative periodontopathogens in Korean advanced adult periodontitis patients using a nucleid acid based approach. J. Periodontol, 71, 1387-1394, 2000.
- 5. Colombo, A.P.; Teles, R.P.; Torres, M.C.; Souto, R.; Rosalem Junior, W.; Mendes, M.C.S.; Uzeda, M. Subgingival microbiota of Brazilian subjects with untreated chronic periodontitis. J. Periodontol., 73, 360-369, 2002.
- 6. Cortelli, S.C.; Jorge, A.O.C.; Cortelli, J.R.; Jordan, S.F.; Haraszthy, V.I. Detection of highly and minimally leukotoxic Actinobacillus actinomycetemcomitans strains in patients with periodontal disease. Pes. Odontol. Bras., 17, 183-188, 2003.
- 7. Cortelli, J.R.; Cortelli, S.C.; Jordan, S.F.; Haraszthy, V.I.; Zambon, J.J. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J. Clin. Periodontol, 32, 860-866, 2005.
- 8. Dahlén, G.; Manji, F.; Baelum, V.; Fejerskov, O. Black pigmented Bacteroides species and Actinobacillus actinomycetemcomitans in subgingival plaque of adult Kenyans. J. Clin. Periodontol, 16, 305-310, 1989.
- 9. Dahlén, G.; Widar, F.; Teanpaisan, R.; Papapanou, P.N.; Baelum, V.; Fejerskov O. Actinobacillus actinomycetemcomitans in rural adult population in southern Thailand. Oral Microbiol. Immunol., 17, 137-142, 2002.
- 10. Dogan, B.; Antinheimo, J.; Cetiner, D.; Bodur, A.; Emingil, G.; Buduneli, E.; Uygur, C.; Firatli, E.; Lakio, L.; Asikainen, S. Subgingival Microflora in Turkish Patients with Periodontitis. J. Periodontol, 74, 803-814, 2003.
- 11. Ellwood, R.; Worthington, H.V.; Cullinan, M.P.; Hamlet, S.; Clerehugh, V.; Davies, R. Prevalence of suspected periodontal pathogens identified using ELISA in adolescents of differing ethnic origins. J. Clin. Periodontol, 24, 141 145, 1997.
- 12. Felmlee, T.; Welch, R.A Alterations of amino acid repeats in the Escherichia coli hemolysin affect cytolytic activity and secretion. Proc. Natl. Acad. Sci. USA, 85, 5273-5279, 1988.
- 13. Fives-Taylor, P.M.; Meyer, D.H.; Mintz, K.P.; Brissette, C. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontol. 2000, 20, 136-167, 1999.
- 14. Flemmig, T.F.; Rudiger, S.; Hofmann, U.; Schmidt, H.; Plaschke, B.; Stratz, A.; Klaiber, B.; Karch, H. Identification of Actinobacillus actinomycetemcomitans in subgingival plaque by PCR. J. Clin. Microbiol., 33, 3102-3105, 1995.
- 15. Haffajee, A.D.; Bogren, A.; Hasturk, H.; Feres, M.; Lopez, N.J.; Socransky, S.S. Subgingival microbiota of chronic periodontitis subjects from different geographic locations. J. Clin. Periodontol, 31, 996-1002, 2004.
- 16. Haubek, D.; Poulsen, K.; Asikainen, S.; Kilian, M. Evidence for absence in northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans J. Clin. Microbiol., 33, 395-401, 1995.
- 17. Haubek, D.; Dirienzo, J.M.; Tinoco, E.M.B.; Westergaard, J.; Lopez, N.J.; Chung, C.P.; Poulsen, K.; Kilian, M. Racial tropism of a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J. Clin. Microbiol., 35, 3037-3042, 1997.
- 18. Haubek, D.; Ennibi, O.K.; Poulsen, K.; Benzarti, N.; Baelum, V. The highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans and progression of periodontal attachment loss. J. Dent. Res., 83, 767-770, 2004.
- 19. Iwase, M.; Lally, T.; Berthold, P.; Korchak, H.M.; Taichman, N.S. Effects of cations and osmotic protetants on cytolytic activity of Actinobacillus actinomycetemcomitans leukotoxin. Infect. Immun., 58, 1782-1788, 1999.
- 20. Lally, E.T.; Golub, E.E.; Kieba, I.R.; Taichman, N.S.; Rosenbloom, J.; Rosenbloom, J.C.; Gibson, C.W.; Demuth, D.R. Analysis of the Actinobacillus actinomycetemcomitans leukotoxin gene. Delineation of unique features and comparison to homologous toxins. J. Biol. Chem., 264, 15451-15456, 1989.
- 21. Lally, E.T.; Golub, E.E.; Kieba, I.R. Identification and immunological characterization of the domain of Actinobacillus actinomycetemcomitans leukotoxin that determines its specificity for human target cells. J. Biol. Chem., 269, 1289-1295, 1989.
- 22. Loesche, W.J.; Lopatin, D.E.; Stoll, J.; van Poperin, N.; Hujoel, P.P. Comparison of various detection methods for periodontopathic bacteria: Can culture be considered the primary reference standard? J. Clin. Microbiol., 30, 418-426, 1992.
- 23. Lopez, N.J. Occurrence of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Prevotella intermedia in progressive adult periodontitis. J. Periodontol, 71, 948-954, 2000.
- 24. Moore, W.E.; Moore, L.V. The bacteria of periodontal diseases. Periodontol. 2000, 5, 66-77, 1994.
- 25. Papapanou, P.N.; Baelum, V.; Luan, W.M.; Madianos, P.N.; Chen, X.; Fejerskov, O.; Dahlén, G. Subgingival microbiota in adult Chinese: prevalence and relation to periodontal disease progression. J. Periodontol, 68, 651-666, 1997.
- 26. Papapanou, P.N.; Teanpaisan, R.; Obiechina, N.S.; Pithpornchaiyakul, W.; Pongpaisal, S.; Pisuithanakan, S.; Baelum, V.; Fejerskov, O.; Dahlen, G. Periodontal microbiota and clinical periodontal status in a rural sample in southern Thailand. Eur. J. Oral Sci., 110, 345-352, 2002.
- 27. Riggio, M.P.; Macfarlane, T.W.; Mackenzie, D.; Lennon, A.; Smith, A.J.; Kinane, D. Comparison of polymerase chain reaction and culture methods for detection Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in subgingival plaque samples. J. Periodontal. Res., 31, 496-501, 1996.
- 28. Slots, J. Rapid identification of important periodontal microorganisms by cultivation. Oral Microbiol. Immunol., 1, 48-55, 1986.
- 29. Slots, J.; Rams, T.E.; Feik, D.; Taveras, H.D.; Gillespie, G.M. Subgingival microflora of advanced periodontitis in the Dominican Republic. J. Periodontol, 62, 543-547, 1991.
- 30. Slots, J.; Ting, M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol. 2000, 20, 82-121, 1999.
- 31. Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.M.; Kent Jr.; R.L. Microbial complexes in subgingival plaque. J. Clin. Periodontol, 25, 134-144, 1998.
- 32. Taichman, N.S.; Simpson, D.I.; Sakurada, S.; Cranfield, M.; DiRienzo, J.M.; Slolts, J. Comparative studies on the biology of Actinobacillus actinomycetemcomitans leukotoxin in primates. Oral Microbiol. Immunol., 2, 97-104, 1987.
- 33. Tan, K.S.; Woo, C.H.; Ong, G.; Song, K.P. Prevalence of Actinobacillus actinomycetemcomitans in an ethnic adult Chinese population. J. Clin. Periodontol., 28, 886-890, 2001.
- 34. Tinoco, E.M.B.; Stevens, R.; Haubek, D.; Lai, C.H.; Balachandran, S.; Preus, H. Relationship of serotype, leukotoxin gene type and lysogeny in Actinobacillus actinomycetemcomitans to periodontal disease status. Eur. J. Oral Sci., 195, 310-317, 1997.
- 35. Tonjum, T.; Haas, R. Identification of Actinobacillus actinomycetemcomitans by leukotoxin gene-specific hybridization and polymerase chain reaction assays. J. Clin. Microbiol., 31, 1856-1856, 1993.
- 36. Umeda, M.; Contreras, A.; Chen, C.; Bakker, I.; Slots, J. The utility of whole saliva to detect oral presence of periodontopathic bactéria. J. Periodontol, 69, 828-833, 1998.
- 37. Yoshida, Y.; Suzuki, N.; Nakano, Y.; Shibuya, K.; Ogawa, Y.; Koga, T. Distribution of Actinobacillus actinomycetemcomitans serotypes and Porphyromonas gingivalis in Japanese adults. Oral Microbiol. Immunol., 18, 135-139, 2003.
- 38. Yuan, K.; Hsu, P-C.; Tseng, C-C.; Kiang, D.; Wang, J-R. Detection rate of Actinobacillus actinomycetemcomitans on the permanent 1st molars of primary school children in Taiwan by polymerase chain reaction. J. Clin. Periodontol, 28, 348-352, 2001.
- 39. Watanabe, K.; Frommel, T.O. Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans and Treponema denticola detection in oral plaque samples using the polymerase chain reaction. J. Clin. Periodontol, 23, 212-219, 1996.
- 40. Willis, S.G.; Smith, K.S.; Dunn, V.L.; Gapter, L.A.; Riviere, K.H.; Riviere, G.R. Identification of seven Treponema species in health- and disease-associated dental plaque by nested PCR. J. Clin. Microbiol., 37, 867-869, 1999.
- 41. Zambon, J.J.; Haraszthy, V.I.; Hariharan, G.; Lally, E.T.; Demuth, D.R. The microbiology of early-onset periodontitis: Association of highly toxic Actinobacillus actinomycetemcomitans strains with localized juvenile periodontitis. J. Periodontol., 67, 282-290, 1996.
Publication Dates
-
Publication in this collection
01 Mar 2007 -
Date of issue
Dec 2006
History
-
Received
01 Sept 2005 -
Reviewed
09 Mar 2006 -
Accepted
18 July 2006