Abstracts
Bacterioplankton was studied in a large area of Southwest Atlantic Ocean between 13 and 25ºS and 28 and 42ºW. Samples were collected in 108 stations at 20 m depth. Bacteria were enumerated by flow cytometry after nucleic acid staining with syto13 and two subgroups were differentiated: low nucleic acid content (LNA) and high nucleic acid content (HNA) bacteria. Total bacterial numbers varied from 0.37 to 5.53 10(5) cells mL-1. HNA cells represented 15 to 70% of the total number while LNA cells represented 30 to 85%. Heterotrophic bacterial production was determined by incorporation of tritiated leucine and ranged from 2.7 to 171.07 ng C L-1 h-1. No significant correlation was found between abundance and production. Nevertheless with support of multivariate analysis between bacterial abundance, bacterial production, chlorophyll a and other oceanographic data the distribution of the groups in two different oceanic provinces could be explained by nutrient availability. HNA bacteria accounted for the high percentage of cells found in the area north of 19ºS, linked to higher temperature waters and riverine nutrients inputs. LNA bacteria were the dominant cells south of this latitude and were correlated to the higher values of nitrate found for the same area.
bacterial abundance; HNA bacteria; LNA bacteria; flow cytometry; Southwest Atlantic Ocean
Um estudo do bacterioplâncton foi realizado numa área extensa do Oceano Atlântico Sudoeste entre 13 e 25ºS e 28 e 42ºW. As amostras foram coletadas em 108 estações oceanográficas a 20 m de profundidade. A abundância bacteriana foi determinada por citometria de fluxo após coloração dos ácidos nucléicos com Syto13. Dois grupos de bactérias foram enumerados e distinguidos: bactérias com alto conteúdo de ácidos nucleicos (HNA) e bactérias com baixo conteúdo de ácidos nucleicos (LNA). O número de bactérias variou de 0,37 a 5,53 10(5) células mL-1. As células HNA representaram de 15 a 70% da abundância total enquanto as células LNA representaram de 30 a 85%. A produção bacteriana foi determinada por incorporação de leucina tritiada e variou de 2,7 a 171,07 ng C L-1 h-1. A correlação entre abundância e produção bacterianas não foi significativa. Entretanto uma análise multivariada realizada entre abundância, produção, clorofila a e outros dados oceanográficos revelou que a distribuição dos dois grupos em diferentes províncias oceânicas pode ser atribuída a disponibilidade de nutrientes. As bactérias HNA foram responsáveis pelo maior percentual de células na área ao norte de 19ºS e estiveram relacionadas às águas quentes e aos nutrientes de origem pluvial. As bactérias LNA foram dominantes ao sul dessa latitude e estiveram relacionadas à disponibilidade de nitrato cujos valores foram mais altos nessa região.
abundância bacteriana; bactérias HNA; bactérias LNA; citometria de fluxo; Oceano Atlântico Sudoeste
ENVIRONMENTAL MICROBIOLOGY
Distribution of HNA and LNA bacterial groups in the Southwest Atlantic Ocean
Distribuição de bactérias HNA e LNA no Oceano Atlântico sudoeste
Luciana AndradeI,* * Corresponding Author. Mailing address: Departamento de Biologia Marinha Instituto de Biologia UFRJ Laboratório de Hidrobiologia Prédio do CCS Bloco A Sala 071 Cidade Universitária Ilha do Fundão cep 21941590 Rio de Janeiro, RJ, Brasil. Tel.: (21) 25626303 ou 22802394. Email: land@biologia.ufrj.br ; Alessandra M. GonzalezI; Carlos Eduardo RezendeII; Marina SuzukiII; Jean Louis ValentinI; Rodolfo ParanhosI
IDepartamento de Biologia Marinha, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
IILaboratório de Ciências Ambientais, Universidade Estadual do Norte Fluminense, Campos dos Goytacazes, RJ, Brasil
ABSTRACT
Bacterioplankton was studied in a large area of Southwest Atlantic Ocean between 13 and 25ºS and 28 and 42ºW. Samples were collected in 108 stations at 20 m depth. Bacteria were enumerated by flow cytometry after nucleic acid staining with syto13 and two subgroups were differentiated: low nucleic acid content (LNA) and high nucleic acid content (HNA) bacteria. Total bacterial numbers varied from 0.37 to 5.53 105 cells mL1. HNA cells represented 15 to 70% of the total number while LNA cells represented 30 to 85%. Heterotrophic bacterial production was determined by incorporation of tritiated leucine and ranged from 2.7 to 171.07 ng C L1 h1. No significant correlation was found between abundance and production. Nevertheless with support of multivariate analysis between bacterial abundance, bacterial production, chlorophyll a and other oceanographic data the distribution of the groups in two different oceanic provinces could be explained by nutrient availability. HNA bacteria accounted for the high percentage of cells found in the area north of 19ºS, linked to higher temperature waters and riverine nutrients inputs. LNA bacteria were the dominant cells south of this latitude and were correlated to the higher values of nitrate found for the same area.
Keywords: bacterial abundance, HNA bacteria, LNA bacteria, flow cytometry, Southwest Atlantic Ocean
RESUMO
Um estudo do bacterioplâncton foi realizado numa área extensa do Oceano Atlântico Sudoeste entre 13 e 25ºS e 28 e 42ºW. As amostras foram coletadas em 108 estações oceanográficas a 20 m de profundidade. A abundância bacteriana foi determinada por citometria de fluxo após coloração dos ácidos nucléicos com Syto13. Dois grupos de bactérias foram enumerados e distinguidos: bactérias com alto conteúdo de ácidos nucleicos (HNA) e bactérias com baixo conteúdo de ácidos nucleicos (LNA). O número de bactérias variou de 0,37 a 5,53 105 células mL1. As células HNA representaram de 15 a 70% da abundância total enquanto as células LNA representaram de 30 a 85%. A produção bacteriana foi determinada por incorporação de leucina tritiada e variou de 2,7 a 171,07 ng C L1 h1. A correlação entre abundância e produção bacterianas não foi significativa. Entretanto uma análise multivariada realizada entre abundância, produção, clorofila a e outros dados oceanográficos revelou que a distribuição dos dois grupos em diferentes províncias oceânicas pode ser atribuída a disponibilidade de nutrientes. As bactérias HNA foram responsáveis pelo maior percentual de células na área ao norte de 19ºS e estiveram relacionadas às águas quentes e aos nutrientes de origem pluvial. As bactérias LNA foram dominantes ao sul dessa latitude e estiveram relacionadas à disponibilidade de nitrato cujos valores foram mais altos nessa região.
Palavraschave:abundância bacteriana, bactérias HNA, bactérias LNA, citometria de fluxo, Oceano Atlântico Sudoeste.
INTRODUCTION
Studies on marine plankton ecology have changed in the last decades to focus on heterotrophic bacteria. Bacteria are players of important roles in the structure and dynamics of food webs and biogeochemical cycles in marine systems (8,21,36,37). Through the microbial loop, dissolved organic carbon (DOC) is transformed into particulated organic carbon (POC) by heterotrophic bacteria and become ready to be incorporated by the next trophic level as these microrganisms are predated. In marine oligotrophic waters this represents more than 50% of POC (3,15). It has also been showed that contribution of heterotrophic bacteria to total plankton biomass and metabolic activity is greater in oligotrophic waters (4,5,9).
Heterotrophic bacteria from water samples can be easily enumerated by flow cytometry, a powerful method available today in aquatic sciences investigation. Combined with cell markers this technique is being increasingly used in environmental microbiology, especially in marine ecosystems (14). The introduction of flow cytometry made bacteria counts faster, easier, and more precise, providing feasibility for experiments involving a high number of counting samples to be processed (22). Also cytometric counts revealed the heterogeneity and differences in bacterioplankton populations allowing the distinction of two general groups: bacteria with high apparent nucleic acid content (HNA) and bacteria with low apparent nucleic acid content (LNA) (1,14,22,26,30,45). The HNA bacteria were considered the most active members of a given community while LNA were regarded as inactive, dead or dormant cells (17,26,27). HNA and LNA were also considered to be members of the same phylotypes (11,38). However some have demonstrated that LNA are different phylotypes from HNA cells (12,31,45) and that indeed LNA can be responsible for a significant portion of total heterotrophic metabolism (1,22,31,34, 45), all observations from oligotrophic systems as open oceans.
The Southwest Atlantic Ocean (SAO) is one of the most oligotrophic areas of the entire oceans, and this can be observed from any chlorophyll satellite image (www.seawifs.gsfc.nasa.gov). Studies on bacteria available for SAO were mostly done with sparse sampling spots taken during transatlantic cruises (20,23,4648). The most recent of these revealed the importance of LNA bacteria in the Atlantic gyres (31). Another study addressed the connection of bacteria abundance and activity to the upwelling phenomena along the SAO (2) and that was the only one referring to a larger oceanographic area. We present here the horizontal distribution of HNA and LNA bacterial subgroups for the same area of the oligotrophic SAO and their relation with the hydrographic data from the area. Our results show that the variable proportions of high and low nucleic acid bacteria differ in oceanic provinces from coastal to open ocean at Brazilian territorial sea.
MATERIALS AND METHODS
Samples were collected between 13 and 25ºS and 28 and 42ºW from April 28th to May 1st, 2000. A grid of 108 oceanographic stations was sampled during the whole cruise from the surface to 200 m depth (Fig. 1). Temperature and salinity profiles were obtained from a CTD Seabird SBE1903 Seacat. The pH was determined on board with an Orion 960 automatic titrator. Dissolved oxygen was determined following Winkler method (CNEXO, 1983) using an automatic titrator (HydroBios 715 Dosimat). Additional physical and chemical data were determined in laboratory based on frozen samples, analyzed in triplicates by standard oceanographical methods (19,35).
Total bacterial abundance was determined after nucleic acid staining with fluorochrome Syto13 at 2.5 µM in samples fixed with paraformaldehyde 2% (10,14). For detection, a FACSCalibur flow cytometer with standard laser and optics was used. For calibration of side scatter and green fluorescence signals, and as an internal standard for cytometric counts and measures, fluorescent latex beads (1.58 µm diameter) were systematically added at a known abundance to each sample (28). The apparent nucleic acid structure was also evaluated, and the results included total bacteria, the bacteria with high apparent nucleic acid content (HNA) and the bacteria with low apparent nucleic acid content (LNA) (see 17, 14 for details).
Bacterial production was determined according to Kirchman et al. (25) and Smith and Azam (40). Triplicates of 1.7 mL of water samples were incubated with 10nM [3H]leucine (Amersham TRK 510, specific activity of 171 Ci nmol1). One tube was amended with 90 µL 100% icecold trichloroacetic acid (TCA) as a control. Incubation was performed at room temperature for 1 h and stopped with 90 µL of 100% icecold TCA. Cells were harvested by centrifugation (13000 rpm, 20ºC, 10 min) and the supernatant was carefully aspirated. Pellets were washed with 1.9 mL of 5% icecold TCA and with 1.7 mL of 80% icecold ethanol, centrifuged and dried as before. After addition of scintillation cocktail, radioactivity was determined in a liquid scintillation counter, with internal calibration and quench correction. Bacterial carbon production was calculated using a protein/carbon conversion factor of 0.86 (41).
Correlations between bacterial and oceanographic data from samples obtained at 20 m depth were determined by Pearson rank correlation index (44). The relationship between all variables was assessed by principal component analysis (29). Data were transformed to logarithmic values to ensure a normal distribution.
RESULTS
Nitrate values ranged from below detection (< 0.05 µM) to 15.89 µM (Table 1), and had a high CV (137%), which can be explained by irregular distribution on the area (Fig. 2a). Values above 5.00 µM were all observed in the southern part of the studied area with the highest value occurring at approximately 23ºS, 40ºW. In this area nitrate varied from 4.37 µM at the surface to 18.12 µM at 200 m, following a nutrient stratification pattern (data not shown). At 20 m depth the highest value of 15.89 µM was found explaining the dark spot on figure 2. The low chlorophyll a values observed (< 0.02 to 1.09 µg L1, average 0.14 µg L1), are characteristic of oligotrophic oceans and are in agreement with values already reported for the area (13,42). The high variability (CV = 99%) explained the differences in distribution (Fig. 2b). The highest values of chlorophyll a were found at 23ºS, 40ºW (where nitrate was also higher) and also associated to riverine inputs.
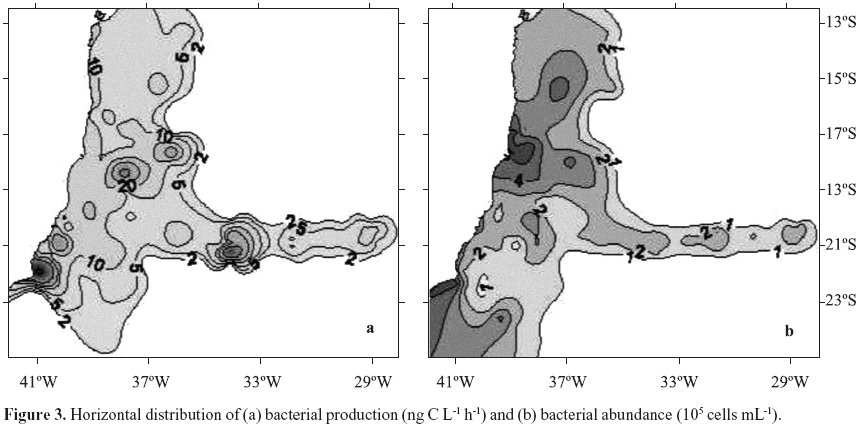
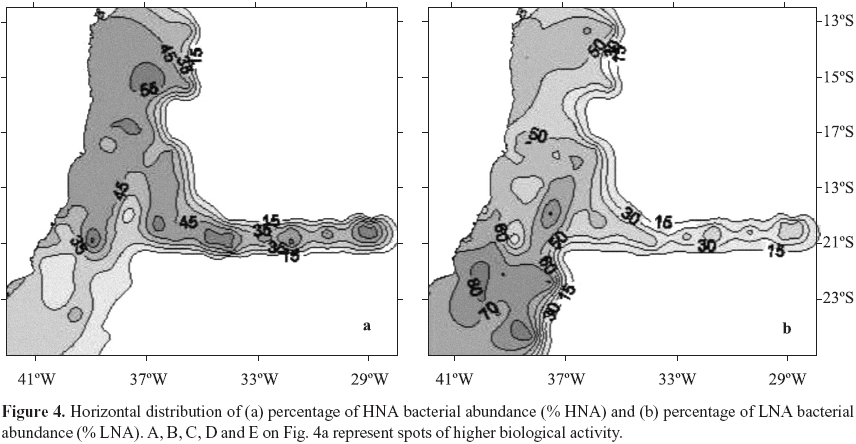
Bacterial production ranged from 2.7 to 171.1 ng C L1 h1, (Table 1). The patchy distribution of bacterial production seen in Fig. 3a is explained by the high variability of these data (CV = 144%). Nuclei of higher values were observed at coordinates 23ºS, 40ºW, Abrolhos Bank and in the beginning of VitóriaTrindade oceanic ridge (around 20ºS; 34ºW). Total bacterial abundance values varied from 0.37 to 5.53 105 cells mL1, CV= 38% (Table 1), values consistent with the ones reported for the Atlantic Ocean (46,48). Although with a low CV (38%) its distribution was heterogeneous. Higher bacterial abundance values occurred in different areas: São Tomé Cape (22ºS, 41ºW), Mucuri river estuary (18ºS, 39º20'W) and Jequitinhonha river estuary (16º20'S, 39ºW) (Fig. 3b). Patches of high values were also found over Abrolhos bank and the oceanic ridge. HNA and LNA abundances were each dominant in different provinces: HNA were more abundant on the northern area of the cruise and over the oceanic ridge and islands (Fig. 4a, above 19ºS) while LNA were dominant mainly in the southern area of the cruise (Fig. 4b, below 19ºS).
The principal component analysis was applied to the matrix of correlation coefficient between 19 variables measured on 107 observations. Figure 5 shows their projection on factorial plan 12 which explains 40.6% of data variability. The most important factor on data variability was attributed to latitudinal gradient (2,13), as the positive coordinates on axis 1 represent the warm and salt water province north of 19ºS and the negative coordinates on axis 1 represent the cold waters south of 19ºS. The distribution of variables confirmed this trend, as south of 19ºS (extreme of negative axis 1) we observed nitrate, N/P ratio and % LNA bacteria with great significance to overall variability. On the positive side of Factor 1 we observed that HNA bacteria and total bacterial abundance were the most important to data distribution on the studied region, followed by nitrogen (except nitrate) and phosphorus.
DISCUSSION
Heterogeneity in distribution of a few parameters was found throughout the SAO (Fig. 1) as confirmed by their high CV's observed (Table 1). This pattern has been observed before (13) and it is explained by the local topography interacting with the complex current flow and water mass structures (39). In distribution maps we identified nuclei of biological activity in the area of the reported São Tomé Eddy (23ºS, 40ºW; ref. 39) mainly represented by nitrate and chlorophyll a (Fig. 2), bacterial activity (Fig. 3a) and the percentage of LNA bacterial abundance (Fig. 4b). Nuclei of biological activity were also found for the VitóriaTrindade ridge (20ºS, 2937ºW) represented by bacterial activity and chlorophyll a (see marks A to E on Fig. 4a). The biological potential of the oceanic ridge was already noticed in punctual observations from 2 different cruises, a decade delayed from each other (2,7). The present data were obtained from a higher number of stations, except for spots B and D (see Fig. 4a) with only one sampling point, so that the patterns obtained can be considered robust and representative of this oceanic feature.
The distribution of bacterial abundance and bacterial production as well as their correlation factors in principal component analysis suggested no coupling between these two variables. This represents a topdown trophic regulation (6) where primary production is dependent on regenerated organic matter from heterotrophic microbial cells. This was already described for the area (2) and it could be a typical pattern in oligotrophic oceans (6). In a given bacterial community not all cells are active, especially in oligotrophic oceans where the resources are scarce and several diverse survival strategies take place (see ref. 32). We were able to discriminate between two types of bacterial cells based on their apparent nucleic acid content (16): the HNA and the LNA groups. However the phylogenetic and functional relevance of cytometrically defined groups still remains poorly understood (31).
HNA bacteria were considered the metabolic active members of a given community, as they were found as responsible for much as 95% of total bacterial carbon production (26). The LNA group has been associated with lower metabolism, inactive, sometimes dormant or even dead cells (16,26). Controversial results were obtained in oligrotrophic ecosystems, where the smaller LNA cells represented the most active members of oceanic systems (22,46) More recently throughout a transect along the Atlantic high aminoacid uptake was attributed to LNA cells which were also the dominant ones in total bacterioplankton numbers (31). We found LNA bacteria dominating the region south of 19ºS (Fig. 4b) where they reached up to 85% of total abundance. The area south 19ºS is characterized by some oceanographic features as eddies and upwelings (13,39,43) that feed the epipelagic zone with cold and nutrientrich waters remobilized from the bottom. This is the most probable explanation for the high nitrate values reported for station 1 and responsible for the dark spot on distribution map (Fig. 2a). In such conditions, the highest growthspecific rates already observed in the smaller LNA cells (45) can explain their relative dominance over bacterial community. The majority of cells in LNA group were already identified as the clade SAR11 of Pelagibacter ubique for the Atlantic Ocean (31). This dominance was predictable due to their small genome size (18). Moreover the SAR11 bacteria were found throughout all oceans accounting for 25 % of all microbial cells (33). Thus these cells could be the active members of bacterial community specialy in oligotrophic waters.
However we also found HNA cells dominating the relative bacterial numbers at waters north of 19ºS (Fig. 4a and 5) which could be attributed to the higher temperature and different nutrient availability as this area is more dependent of rivers discharges. Nowadays a myriad of metabolic states are recognized for aquatic bacteria (22), and which parcel of population is responsible for the bulk of activity is still a controversy. SAO lacks comprehensive research on microbial food web structures and function. Constant monitoring probably holds the key for understanding the oceanic carbon cycle and its interdependence on climate change (24). Further future research in the area should try to address the changes in bacterioplankton metabolism and diversity across trophic gradients in open ocean.
ACKNOWLEDGEMENTS
Thanks are due to the officers and crew of the R.V. Astro Garoupa; Environmental Ministry and PETROBRAS for ship time and fuel. We thank F. Matos, R. Guimarães, M. Semeraro, C. Trisziucci, A. Reis, G. Oliveira, J. Siqueira, N. Holmes, and J.O. Nascimento for help in field work. We are grateful to Dr. M. Bellio and Dr. A. Nóbrega, (Dept. Imunology, UFRJ) for the friendly access to flow cytometer, and to Prof. F.A. Esteves (Lab. Limonology, UFRJ) for the use of LSC. This research was supported by Brazilian Environment Ministry and SeCIRM, PRONEX O887/96, CNPq, FAPERJ E26/171.265/97 and FUJBUFRJ 40967.
Submitted: August 08, 2005; Returned to authors for corrections: January 18, 2006; Approved: February 23, 2007.
- 1. Andrade, L.; Gonzalez, A.M.; Araujo, F.V.; Paranhos, R. (2003). Flow cytometry assessment of bacterioplankton in tropical marine environments. J. Microbiol. Meth, 55, 841850.
- 2. Andrade, L.; Gonzalez, A.M.; Valentin, J.L.; Paranhos, R. (2004). Bacterial abundance and production in the southwest Atlantic Ocean. Hydrobiologia, 511, 103111.
- 3. Azam F.; Cho, B.C. (1987). Bacterial utilization of organic matter in the sea. In: Fletcher M., T.R.G. Gray & J.G. Jones (eds). Ecology of microbial communities, Cambridge University Press, p.261281.
- 4. Biddanda, B.A.; Opsahl S.; Benner R. (1994). Plankton respiration and carbon flux through bacterioplankton on the Louisiana shelf. Limnol. Oceanogr, 39, 12591275.
- 5. Biddanda, B.A.; Ogdahl M.; Cotner J. (2001). Dominance of bacterial metabolism in oligotrophic relative to eutrophic waters. Limnol. Oceanogr, 46(3) 730739.
- 6. Billen, G.; Servais, P.; Becquevort, S. (1990). Dynamics of bacterioplankton in oligotrophic and eutrophic aquatic environments: Bottomup and topdown control? Hydrobiologia, 207, 3742.
- 7. Bonecker, S.L.; Nogueira, C.R.; Bonecker, A.C.T.; Santos, L.H.S.; Reynier, M.; Tenenbaum, D.R. (1992/93). Estudo hidrográfico e planctonológico da região entre Cabo Frio (RJ) e o arquipélago de Abrolhos (BA), Brasil. Neritica, 7(12) 7186.
- 8. Cole J.J.; Findlay S.; Pace M.L. (1988). Bacterial production in fresh and saltwater ecosystems: a crosssystem overview. Mar. Ecol. Prog. Ser, 43, 110.
- 9. Del Giorgio P.; Cimbleris A. (1997). Respiration rates in bacteria exceed phytoplankton production in unproductive aquatic systems. Nature, 385, 148151.
- 10. Del Giorgio, P.A.; Bird, D.F.; Prairie, Y.T.; Planas, D. (1996). Flow cytometric determination of bacterial abundance in lake plankton with the green stain SYTO 13. Limnol. Oceanogr, 41, 11691179.
- 11. Flaten, G.A.F.; Castberg, T.; Tanaka, T.; Thingstad, T.F. (2003). Interpretation of nutrientenrichment bioassays by looking at subpopulations in a marine bacterial community. Aquat. Microb. Ecol, 33, 1118.
- 12. Fuchs, B.M.; Woebken, D.; Zubkov, M.V.; Burkill, P.; Amann, R. (2005). Molecular identification of picoplankton populations in contrasting waters of the Arabian Sea. Aquat. Microb. Ecol, 39, 145157.
- 13. Gaeta, S.A.; Lorenzetti, S.A.; Miranda, L.B.; SusiniRibeiro, S.M.M.; Pompeu, M.; Araújo, C.E.S. (1999). The Vitória Eddy and its relation to the phytoplankton biomass and primary productivity during the austral fall of 1995. Arch. Fish. Mar. Res., 47, 253270.
- 14. Gasol, J.M.; del Giorgio, P.A. (2000). Effects of filtration on bacterial activity and picoplankton community structure as assessed by flow cytometry. Aquatic. Microb. Ecol, 16, 251264.
- 15. Gasol, J.M.; Duarte, C.M. (2000). Comparative analyses in aquatic microbial ecology: how far do they go? FEMS Microb. Ecol, 31, 99106.
- 16. Gasol J.M.; Doval, M.D.; Pinhassi, J.; CalderónPaz, J.I.; GuixaBoixareu, N.; Vaqué D.; PedrósAliós, C. (1998). Diel variations in bacterial heterotrophic activity and in the northwestern Mediterranean Sea. Mar. Ecol. Prog. Ser, 164, 107124.
- 17. Gasol, J.M.; Zweifel, UL.; Peters, F.; Furhman, J.A.; Hagström, Å. (1999). Significance of size and nucleic acid content heterogeneity as assessed by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol, 65, 44754483.
- 18. Giovannoni, S.J.; Tripp, H.J.; Givan, S.; Podar, M. and 10 others. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science, 309, 12421245.
- 19. Grasshoff, K.A.; Ehrhardt, M.; Kremling, K. (1983). Methods of Seawater Analysis 2nd ed. Verlag Chemie, 419p.
- 20. Heywood, J.L.; Zubkov, M.V.; Tarran, G.A.; Fuchs, B.M.; Holligan, P.M. (2006). Prokarioplankton standing stocks in oligotrophic gyre and equatorial provinces of the Atlantic Ocean: evaluation of interannual variability. DeepSea Res. II, 53, 15301547.
- 21. Hoppe H.G.; Giesenhagen H.C.; Gocke, K. (1998). Changing patterns of bacterial substrate decomposition in an eutrophication gradient. Aquat. Microb. Ecol., 15, 113.
- 22. Jochem, F.J.; Lavrentyev; P.J.; First, M.R. (2004). Growth and grazing rates of bacteria groups with different apparent DNA content in the Gulf of Mexico. Mar. Biol., 145, 12131225.
- 23. Jürgens, K.; Gasol, J.M.; Vaqué, D. (2000). Bacteriaflagellate coupling in microcosm experiments in the Central Atlantic Ocean. J. Exp. Mar. Biol. Ecol., 245, 127147.
- 24. Karl, D.M. (2004). Microbial biogeochemistry in the north pacific subtropical gyre. 10th International Conference on Microbial Ecology ISME10, p.111.
- 25. Kirchman, D.L.; K'nees, E.; Hodson, R. (1985). Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol., 49, 599607.
- 26. Lebaron, P.; Servais, P.; Agongué, H.; Courties, C.; Joux, F. (2001). Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol., 67, 17751782.
- 27. Lebaron, P.; Servais, P.; Baudoux, AC.; Bourrain, M.; Courties, C.; Parthuisot, N. (2002). Variations of bacterialspecific activity with cell size and nucleic acid content assessed by flow cytometry. Aquatic Microbial Ecology, 28, 131140.
- 28. Lebaron, P.; Troussellier, M.; Got, P. (1994). Accuracy and precision of epifluorescence microscopy counts for direct estimates of bacterial numbers. J. Microbiol. Methods, 19, 8994.
- 29. Légendre, L.; Légendre, P. (1998). Écologie numérique Tome 2: La structure dês donnés écologique. Collection d'Écologie, 13. Masson, Paris et les Presses de l'Université du Québec.
- 30. Li, W.K.W.; Jellett, J.F.; Dickie, P.M. (1995). DNA distribution in planktonic bacteria stained with TOTO or TOPRO. Limnol. Oceanogr., 40, 14851495.
- 31. Mary, I.; Heywood, J.L.; Fuchs, B.M.; Amann, R.; Burkill, P.H.; Tarran, G.A.; Zubkov, M.V. (2006). SAR11 dominance among metabolically active low nucleic acid bacterioplankton in surface waters along an Atlantic meridional transect. Aquat. Microb. Ecol., 45, 107113.
- 32. Morita, R.Y., (1997). Bacteria in Oligotrophic Environments. StarvationSurvival Lifestyle Chapman e Hall, New York, 608p.
- 33. Morris, R.M.; Rappe, M.S.; Connon, S.A.; Vergin, K.L.; Siebold, W.A.; Carlson, C.A.; Giovannoni, S.G. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature, 420, 806810.
- 34. Nishimura, Y.; Kim, C.; Nagata, T. (2005). Vertical and seasonal variations of bacterioplankton subgroups with different nucleic acid contents: possible regulation by phosphorus. Appl. Environ. Microbiol, 71, 58285836.
- 35. Parsons, T.R.; Maita, Y.; Lalli, C.M. (1984). A Manual of Chemical and Biological Methods for Seawater Analysis Pergamon Press, Oxford, pp.173
- 36. Schauer M.; Massana R.; PedrósAliós, C. (2000). Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol, 33, 5159.
- 37. Servais P. (1992). Bacterial production measured by 3Hthymidine and 3Hleucine incorporation in various aquatic ecosystems. Arch. Hydrobiol. Beih. Ergebn. Limnol., 37, 7381.
- 38. Servais, P.; Casamayor, E.O.; Courties, C.; Catalla, P.; Parthuisot, N.; Lebaron, P. (2003). Activity and diversity of bacterial cells with high and low nucleic acid content. Aquat. Microb. Ecol, 33, 4151.
- 39. Silveira, I.C.A.; Calado, L.; Castro, B.M.; Cirano, M.; Lima, J.A.M.; Mascarenhas, A S. (2004). On the baroclinic strucutre of the Brazil CurrentIntermediate Western Boundary Current system at 22º23ºS. Geophys. Res. Lett, 31, LI4308, 15.
- 40. Smith, D.C.; Azam, F. (1992). A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3Hleucine. Mar. Microbial Food Webs, 6, 107114.
- 41. Simon, M.; Azam, F. (1989). Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser., 51, 201213.
- 42. SusiniRibeiro, S.M.M. (1999). Biomass distribution of pico, nano and microplankton on the continental shelf of Abrolhos, East Brazil. Arch. Fish. Mar. Res, 47, 271284.
- 43. Valentin, J.L.; André, D.L.; Jacob, A.S. (1987). Hydrobiology in the Cabo Frio (Brazil) upwelling: twodimensional structure and variability during a wind cycle. Cont. Shelf Res., 7: 7788.
- 44. Zar, J.H. (1984). Biostatistical Analysis 2nd ed. PrenticeHall, New Jersey.
- 45. Zubkov, M.V.; Fuchs, B.M.; Burkill, P.H.; Amann, R. (2001a). Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol., 67, 52105218.
- 46. Zubkov, M.V.; Sleigh, M.A.; Burkill, P.H. (2000). Assaying picoplankton distribution by flow cytometry of underway samples collected along a meridional transect across the Atlantic Ocean. Aquatic. Microb. Ecol., 21, 1320.
- 47. Zubkov, M.V.; Sleigh, M.A.; Burkill, P.H. (2001b). Heterotrophic bacterial turnover along the 20ºW meridian between 59ºN and 37ºN in July 1996. DeepSea Res II, 48, 9871001.
- 48. Zubkov, M.V.; Sleigh, M.A.; Tarran, G.A.; Burkill, P.H.; Leakey, R.J.G. (1998). Picoplankton community structure on an Atlantic transect from 50ºN to 50ºS. DeepSea Res. I, 45, 13391355.
Publication Dates
-
Publication in this collection
29 June 2007 -
Date of issue
June 2007
History
-
Accepted
23 Feb 2007 -
Reviewed
18 Jan 2006 -
Received
08 Aug 2005