Abstracts
Archaea represent a considerable fraction of the prokaryotic world in marine and terrestrial ecosystems, indicating that organisms from this domain might have a large impact on global energy cycles. The extremophilic nature of many archaea has stimulated intense efforts to understand the physiological adaptations for living in extreme environments. Their unusual properties make them a potentially valuable resource in the development of novel biotechnological processes and industrial applications as new pharmaceuticals, cosmetics, nutritional supplements, molecular probes, enzymes, and fine chemicals. In the present mini-review, we show and discuss some exclusive characteristics of Archaea domain and the current knowledge about the biotechnological uses of the archaeal enzymes. The topics are: archaeal characteristics, phylogenetic division, biotechnological applications, isolation and cultivation of new microbes, achievements in genomics, and metagenomic.
Archaea; Biotechnology; Extremozymes; Genomic; Molecular Phylogeny
As arqueas representam uma considerável fração dos procariotos nos ecossistemas marinhos e terrestes, indicando que estes organismos devem possuir um grande impacto nos ciclos energéticos. A natureza extremofílica de muitas arqueas tem estimulado intensos esforços para compreender sua adaptação fisiológica a ambientes extremos. Suas propriedades incomus as tornam uma fonte valiosa no desenvolvimento de novos processos biotecnológicos e aplicações industriais como novos fármacos, cosméticos, suplementos nutricionais, sondas moleculares, enzimas e reagentes. Na presente mini-revisão, mostramos e discutimos algumas de suas características exclusivas correlacionando-as com seu potencial biotecnológico e aplicação industrial. Os tópicos são: características das arqueas, divisão filogenética, aplicações biotecnológicas, isolamento e cultivo de novos microrganismos, genoma e metagenoma.
Archaea; Biotecnologia; Enzimas extremofílicas; Filogenia Molecular; Genômica
MINI-REVIEW MICROBIOLOGY
MINI-REVIEW
Exploring the biotechnologial applications in the archaeal domain
Explorando as aplicações biotecnológicas do domínio archaea
Alquéres, S.M.C.I; Almeida, R.V.II; Clementino, M.M.III; Vieira, R.P.I; Almeida, W.I.I; Cardoso, A.M.I,* * Corresponding Author. Mailing Address: Instituto de Bioquímica Médica, Universidade Federal do Rio de Janeiro, Centro de Ciências da Saúde - Bloco D, subsolo, sala 5, 21941-590 - Rio de Janeiro - Brasil. Tel.: +55 21 25626751 - Fax: +55 21 22708647. E-mail: amcardoso@bioqmed.ufrj.br ; Martins, O.B.I
IInstituto de Bioquímica Médica, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
IIInstituto de Química, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
IIIInstituto Nacional de Controle da Qualidade em Saúde, Fundação Instituto Oswaldo Cruz, Rio de Janeiro, RJ, Brasil
ABSTRACT
Archaea represent a considerable fraction of the prokaryotic world in marine and terrestrial ecosystems, indicating that organisms from this domain might have a large impact on global energy cycles. The extremophilic nature of many archaea has stimulated intense efforts to understand the physiological adaptations for living in extreme environments. Their unusual properties make them a potentially valuable resource in the development of novel biotechnological processes and industrial applications as new pharmaceuticals, cosmetics, nutritional supplements, molecular probes, enzymes, and fine chemicals. In the present mini-review, we show and discuss some exclusive characteristics of Archaea domain and the current knowledge about the biotechnological uses of the archaeal enzymes. The topics are: archaeal characteristics, phylogenetic division, biotechnological applications, isolation and cultivation of new microbes, achievements in genomics, and metagenomic.
Key-words: Archaea, Biotechnology, Extremozymes, Genomic, Molecular Phylogeny.
RESUMO
As arqueas representam uma considerável fração dos procariotos nos ecossistemas marinhos e terrestes, indicando que estes organismos devem possuir um grande impacto nos ciclos energéticos. A natureza extremofílica de muitas arqueas tem estimulado intensos esforços para compreender sua adaptação fisiológica a ambientes extremos. Suas propriedades incomus as tornam uma fonte valiosa no desenvolvimento de novos processos biotecnológicos e aplicações industriais como novos fármacos, cosméticos, suplementos nutricionais, sondas moleculares, enzimas e reagentes. Na presente mini-revisão, mostramos e discutimos algumas de suas características exclusivas correlacionando-as com seu potencial biotecnológico e aplicação industrial. Os tópicos são: características das arqueas, divisão filogenética, aplicações biotecnológicas, isolamento e cultivo de novos microrganismos, genoma e metagenoma.
Palavras-chave: Archaea, Biotecnologia, Enzimas extremofílicas, Filogenia Molecular, Genômica.
Archaea domain
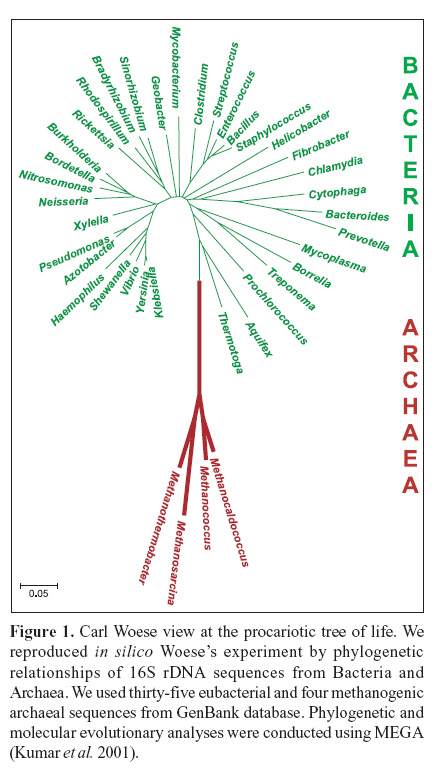
In the late 1970s, Carl Woese and his colleagues at the University of Illinois studied relationships among prokaryotes and proposed that life should be divided into three domains: Archaea, Bacteria, and Eucarya. They found that there were two distinctly different prokaryotic groups. Those "bacteria" that lived at high temperatures or produced methane were clustered together as a group far away from the usual bacteria (Fig. 1) and the eukaryotes (51).
Woese argued that Archaea, Bacteria, and Eucarya each represent a primary line of descent that diverged early from an ancestral progenote with poorly developed genetic machinery. This hypothesis is reflected in the name Archaea, from the Greek archae, meaning ancient. Later he treated these groups formally as domains, each consisting of several kingdoms. This separation and organization of tree of life have become very popular, although the idea of the progenote itself is not generally supported (50).
For 15 years after their recognition, the Archaea were generally known only as inhabiting hostile environments (27). Within the past two decades, the use of molecular techniques, including PCR-based amplification of 16S rRNA genes, has allowed a culture-independent assessment of microbial diversity, indicating a wide distribution of mostly uncultured archaea in normal habitats, such as ocean and lake waters and soil (10). Nowadays the use of 16S rDNA clone libraries to map the diversity of uncultivated prokaryotes from natural populations has provided a revolutionary advance for interpreting microbial evolutionary relationships. These molecular surveys have produced more than 20,000 archaeal 16S rRNA gene sequences from environmental studies, extending the known groups and increasing the number of novel lineages. In Brazil, our group study the archaeal diversity in tropical environments and show that the detection of a substantial number of uncultured phylotypes suggests that this region harbors a pool of novel archaeaplankton taxa (11,47).
Archaeal characteristics
Several features set the Archaea apart, for example, archaea have a single cell membrane containing a peptidoglycan-like wall which is different in bacteria. Furthermore, both eubacterias and eukaryotes have membranes composed mainly of glycerol-ester lipids, whereas archaea have membranes composed of glycerol-ether lipids. These differences may be an adaptation to extreme environments (7). Archaeal organisms also have flagella that are notably different in composition and development from the flagella of bacteria. Individual archaea cells range from 0.1 to over 15 µm in diameter, and some form aggregates or filaments up to 200 µm in length. They occur in various shapes, such as spherical, rod-shaped, spiral, lobed, or rectangular, and they also exhibit a variety of different types of metabolism. Archaeal and bacterial metabolic genes share common evolutionary aspects (6). However, the transcriptional and translational machinary of Archaea is much more similar to Eucarya than Bacteria (28). For instance, archaean translation uses eukaryotic initiation and elongation factors, and their transcription involves TATA-binding proteins and TFIIB (14). The study of protein synthesis in Archaea led to a much deeper understanding of this process not only in the Archaea, but in all domains of life (8,33,52).
Many archaeans are extremophiles, some live at very high temperatures, often above 100ºC, as found in geysers and submarine black smokers. Others are found in very cold habitats, highly saline, acidic, or alkaline water. They are able to live in the anoxic mud of marshes and at the bottom of the ocean, even thrive in petroleum deposits deep underground. However, many archaeans are mesophiles, and have been found in environments like marine plankton (5), sewage, and soil (36). Futhermore, many methanogenic archaea are found in the digestive tracts of animals such as ruminants, termites, and humans (15,42).
Phylogenetic division
The Archaea domain is divided on the basis of 16S rRNA gene sequences into four phylogenetically distinct phylum: Euryarchaeota, Crenarchaeota, Korarchaeota and Nanoarchaeota (Fig. 2).
Cultivated crenarchaeotes presents a thermoacidophilic phenotype. The term "cren" means spring or fount and expresses the resemblance of this phenotype to the ancestor of the domain Archaea. Most cultured representatives of the Crenarchaeota are extreme thermophiles or hyperthermophiles. A broad variety of metabolic pathways is evident. Aerobically growing chemolithotrophs gain energy by the oxidation of various sulfur compounds, molecular hydrogen or ferrous iron. Anaerobic chemolithotrophs reduce sulfur, thiosulfate or produce nitrate, hydrogen sulfide or ammonia. Organotrophic growth occurs on complex organic substrates, sugars, amino acids or polymers such as starch and cellulose. However, cultivated species appear to represent a minority in Crenarchaeota, which comprise an important number of uncultured groups from marine plankton, freshwater, and soil samples (13,33,39)
The Euryarchaeota phylum, the term "eury" means broad or wide, contains organisms that are highly diverse in their physiology, morphology and natural habitats. During the last decade, numerous reclassifications within the Euryarchaeota have been carried out, mainly based on the results of 16S rRNA sequence comparisons. Five major groups are known within this kingdom: the obligate anaerobic methanogens, the extreme halophiles, the hyperthermophilic sulfate reducers, the Thermoplasma group, and finally, the Thermococcus-Pyrococcus group (28, 35).
Interesting, new microbes are still being discovered and investigated. Most recently, Karl Stetter and his colleagues discovered the world's smallest microorganism, Nanoarchaeum equitans. Its name roughly translates to "ancient dwarf who rides the fire ball" because of its tendency to latch on to or "ride" the surface of the archaeal microbe Ignicoccus ("fireball"). The discovery of this nanosized hyperthermophilic archaeon has led to the creation of a new phylum, Nanoarchaeota (19). This extremophile was found in a 120 m depth submarine hydrothermal vent and thrives in temperatures close to 100ºC. With less than 500 kb in its genome, N. equitans represents the smallest archaeal genome sequenced until now.
Recently, Korarchaeota-specific primers were designed based on previously published sequences and used to screen a variety of environments. Korarchaeota 16S rRNA genes were amplified from high temperature like Yellowstone National Park hot springs, East Pacific Rise deep-sea hydrothermal vent and also found in other environments (3).
Although this is a well established division, some authors have suggested a revision in this classification as sequences from environmental samples are emerging, showing that the archaeal tree is more complex than expected (35).
Biotechnological Applications
The extremophilic nature of many Archaea has stimulated intense efforts to understand the physiological adaptations for living in extremes environments and to probe the potential biotechnological applications of their stable cellular components. This is particularly true of their enzymes (called extremozymes), which are able to remain catalytically active under extremes of temperature, salinity, pH and pressure. Many interesting enzymes have been isolated from extremophilic microbes (41). Specific archaeal metabolites have also been purified and characterized and some of them have potential industrial uses (Table 1). However, several technical difficulties have prevented the large-scale industrial application of enzymes and special metabolites from extremophilic sources, the most important being the availability of these compounds (40). Genes encoding several enzymes from extremophiles have been cloned in mesophilic hosts, with the objective of overproducing the enzyme and altering its properties to suit commercial applications. Escherichia coli,Bacillus subtilis and yeasts have been used successfully as mesophilic hosts for several archaeal genes. Genetic engineering techniques are valuable tools for creating novel biocatalysts that can improve bioprocesses and facilitate the realization of innovative biotransformations. Molecular biology has the potential both to overcome the limitations on enzyme availability and to design new biocatalysts specific to a particular industrial purposes.
Gluco hydrolyzing enzymes
Due to the complex structure of starch, a number of enzymes are needed for its degradation. Endo-acting enzymes, such as a-amylase, hydrolyse linkages in the interior of the starch polymer in a random fashion, which leads to the formation of linear and branched oligosaccharides. Exo-acting enzymes (a and b-glucoamylases) attack the substrate from the non-reducing end, producing oligo and/or monosaccharides. The finding of new extremely thermostable starch-hydrolysing enzymes, such as amylases and pullulanases, will significantly improve the industrial starch bioconversion process. Investigations concerning recombinant a-amylases from Pyrococcus woesei indicate their suitability for starch processing. Furthermore, the study of recombinant b-galactosidase from P. woesei suitable for purpose of low lactose milk and whey production is also presented. The activity of this enzyme in a wide pH range of 4.3-6.6 and high thermostability suggests that it can be used for processing of dairy products at temperatures which restrict microbial growth during a long operating time of continuous-flow reactor with an immobilized enzyme system. Preparation of recombinant a-amylase and b-galactosidase was facilitated by cloning and expression of genes from P. woesei in E. coli host. Satisfactory level of recombinant enzymes purification was achieved by thermal denaturation and precipitation of native proteins originated from E. coli. The obtained a-amylase has maximal activity at pH 5.6 and 93ºC. The half-life of this preparation (pH 5.6) at 90ºC and 110ºC was 11 h and 3.5 h, respectively, and retained 24% of residual activity following incubation for 2 h at 120ºC (46).
A hyperthermostable glycosidase enzyme with pullulanase activity at 90ºC from Thermococcus aggregans was cloned and expressed in E. coli. Unlike all other pullulan-hydrolyzing enzymes described, the enzyme is able to attack a-1,6- as well as a-1,4-glycosidic linkages, affording a mixture of maltotriose, maltose and glucose. The enzyme is also able to degrade starch, amylose and amylopectin, forming maltotriose and maltose as main products (26).
Halophilic glycosidases have also been identified, including a halophilic b galactosidase from Haloferax alicantei. The extremely halophilic galactosidase from Haloferax alicantei is optimally active at 4 M NaCl. Purification of the enzyme was facilitated by the ability of sorbitol to stabilize enzyme activity in the absence of salt, which allowed conventional ion-exchange chromatography (18).
The continuous production of halophilic a-amylase can be performed via whole-cell immobilization of Halobacterium salinarum in alginate beads and a polyvinyl alcohol film. The cells were osmotically stable and showed continuous enzyme production for 45 days. The stabilized cells could be permeabilized by chloroform treatment without leakage of the intracellular components. Using this procedure, cells can be reused under improved stabilized conditions for biotechnological applications (4).
Proteases and Peptidases
The amount of proteolytic enzymes produced world-wide on a commercial scale is larger than that of any of the other biotechnologically used enzymes. Serine alkaline proteases are used as additives to household detergents for laundering where they must resist denaturation by detergents and alkaline conditions. The leather industry uses proteinases with high keratinolytic and elastolytic activities for soaking. The discovery of proteases that can catalyze reactions under extreme conditions (high temperatures and extremes of pH) will be valuable for industrial applications (23,25).
An extracellular serine protease from the extreme halophile Halobacterium halobium may be an excellent catalyst for peptide synthesis, exploiting their reverse reaction, particularly for glycine-containing peptides. The enzyme requires 4 M NaCl for optimal catalytic activity and stability in aqueous solutions (38). The stabilization of halophilic enzymes by organic solvents while lowering the required salt concentration is of practical importance because high NaCl concentrations can be corrosive to metals. Higher substrate solubility in the presence of an organic solvent can be useful in synthetic reactions catalyzed by enzymes from extreme halophiles (21). Many biotechnologically interesting serine proteases have been identified and characterized. Among them is a cell associated serine protease, characterized from Desulfurococcus strain SY, which showed a half-life of 4.3 h at 95ºC (17). A globular serine protease from Staphylothermus marinus was found to be extremely thermostable. This enzyme has residual activity even after 10 min of incubation at 135ºC (24). Another gene encoding a subtilisin-like serine protease, named aereolysin, has been cloned from Pyrobaculum aerophilum and the protein was modelled on the basis of structures of subtilisin-type proteases (48). Multiple proteolytic activities have been observed in Pyrococcus furiosus. The cell envelope associated serine protease of P. furiosus, called pyrolysin, was found to be highly stable with a half-life of 20 min at 105ºC. The pyrolysin gene was cloned and sequenced and it was shown that this enzyme is a subtilisin-like serine protease (25,49).
Proteases have also been characterized from the thermoacidophilic archaeon Sulfolobus solfataricus and Sulfolobus acidocaldarius. In addition to the serine proteases, other types of enzymes have been identified in extremophiles. Three good examples are a thiol protease (propylpeptidase) from Pyrococcus sp. and a new type of protease from P. furiosus (25).
Psychrophilic enzymes have potential applications in a broad range of industrial, agricultural and medical processes. The recent interest in this field is due to the challenge of finding stable proteases that function in cold water. In addition, peptidyl synthesis studies with mesophilic enzymes have shown that low temperature favors high yields because of reduced hydrolysis of the acyl-enzyme intermediate. In most other processes, reduced energy consumption due to low temperature operation will be a significant advantage (9).
DNA processing enzymes
One of the most important advances in molecular biology was the development of the polymerase chain reaction (PCR). Thermostable DNA polymerases play a major role in PCR and in a variety of molecular biological applications, e.g. DNA amplification, cloning, sequencing or labeling. Several native and recombinant polymerases have been purified and characterized (32).
DNA polymerase I from the bacterium Thermus aquaticus, called Taq polymerase, was the first thermostable DNA polymerases utilized in PCR. Archaeal proof-reading polymerases, such as Pwo from Pyrococcus woesei,Pfu from P. furiosus, Deep Vent polymerase from the Pyrococcus strain GB-D and Vent polymerase from Thermococcus litoralis, have an error rate that is up to tenfold lower than that of Taq polymerase (44).
Esterases and lipases
In the field of biotechnology, esterases are receiving increasing attention because of their application in organic biosynthesis. In aqueous solution, esterases catalyze the hydrolytic cleavage of esters to form the constituent acid and alcohol whereas, in organic solutions, the transesterification reaction is promoted. Both the reactants and the products of transesterification are usually highly soluble in the organic phase and the reactants may even form the organic phase themselves. The P. furiosus esterase and lipase (1) genes have been cloned in E. coli and the functional properties have been determined. The archaeal enzyme is the most thermostable (a half-life of 50 min at 126ºC) and thermoactive (optimum temperature of 100ºC) esterase known to date (20).
Archaeosomes
Liposomes are lipid-bilayer bounded vesicles that can be used as a delivery vehicle for certain vaccines, enzymes, and drugs. The term "archaeosome" was introduced by Sprott and co-workers to describe liposomes made with ether lipids that are unique to the Archaea domain confering considerable stability on liposomal vesicles (45). Extensive mouse model studies involving intravenous, oral and subcutaneous administration of archaeosomes demonstrated that archaeosomes are safety molecules and they are not toxic (31).
Archaeal S-layer
Like other elements of the cell envelope, S-layers play an important part in interactions of the microbial cell with the environment (43). It was demonstrated that archaeal S-layers have been shown to be excellent patterning structures in molecular nanotechnology and biomineralization due to their high molecular order, high binding capacity and ability to recrystallize with perfect uniformity on solid surfaces, at the air:water interface and on lipid films (43).
Bacteriorhodopsin
The halophilic archaeon makes use of light for both energy and sensory transduction by exploiting a family of light-sensitive proteins. The archaeal rhodopsin, a 26.5 kDa protein, has a transmembrane domain of seven helical protein segments which photons are captured via a retinal chromophore (29). The excellent thermodynamic and photochemical stability of bacteriorhodopsin has led to many uses in technical applications like holography, spatial light modulators, artificial retina, neural network optical computing, and volumetric and associative optical memories (22).
Isolation and cultivation of new microbes
Several studies specifically aimed at the isolation of archaeal DNA rather than at the isolation of the microorganisms themselves are known (13,30,39). However, the isolation of microorganisms from complex mixed cultures and their cultivation in a pure culture is an essential prerequisite for their complete characterization. Different processes are used for this purpose, including the classic plate casting process, dilution methods, and selective enrichment. The majority of cells obtained from nature do not generally form visible colonies on plates (16). It can be very difficult to isolate new microbes when the conditions for their growth are not known or when the microbes grow in a heterogeneous mixed culture in very small numbers (12). Evidently, the biosphere remains replete with unexplored niches filled with extraordinary organisms awaiting the design of new cultivation and isolation strategies (34).
Genomics
Many microorganisms have been isolated from the Archaea domain, and among the archaeal enzymes that have been purified and characterized, several show potential for industrial use. Archaeal metabolites suitable for industrial applications have also been found (Table 1). In addition to the development of industrial production processes, basic research is also needed to determine how enzymes have adapted to extreme conditions at the molecular level. It has been shown that modifications in enzymes of psychrophiles to permit conformational flexibility at low temperatures are opposite to those that confer thermostability to proteins of thermophiles (37). The slow incorporation of archaeal tools by industry can be ascribed mainly to the difficulties associated with the development of large-scale processes for the production of archaeal biomasses.
Genome sequencing should help to overcome these difficulties. Cloning into an easy-to-grow mesophilic host may be the preferred method for archaeal enzyme production because it simplifies downstream processing. Strategies that combine molecular biology techniques and customized bioprocess design could greatly improve the yields of archaeal bioproducts. This would result in major cost savings and therefore, enable industrial applications of these unique biomaterials.
Metagenomics: searching for new genes
Molecular studies of microbial diversity have shown that not cultivated microorganisms can correspond to >99% of the total microbial population (2). Current knowledge of prokaryotic diversity is so poor, in fact, that a significant number of archaea isolated randomly from natural environments will turn out to be a new organism. Furthermore, access to this reservoir of genetic and metabolic diversity is of great interest in biotechnology. The main requirement is the availability of pure, intact, high molecular weight genomic DNA. High-molecular-weight DNA extracted from environmental samples can be cloned directly into large insert cloning vectors, such as cosmids, fosmids, or bacterial artificial chromosomes (BACs) (36). The genome fragments of uncultivated microbes can useful for expressing protein-encoding genes, determining enzyme structure and function, or dissecting metabolic pathways. Examples of such an approach resulting in the genomic analysis are the construction of large-insert bacterial artificial chromosome (BAC) libraries from the genomic DNA of planktonic marine microbial assemblages (5) and from genomic DNA isolated directly from soil (36).
Perspectives
Many microorganisms have been isolated from the domain Archaea, and among the archaeal enzymes that have been purified and characterized, several show potential for industrial uses. However, biotechnological processes based on these archaeal tools remain limited, despite the extensive recent literature suggesting possible novel bioprocesses and demonstrating the applicability of several biomolecules of archaeal origin in the biotechnology industry. Molecular biology has the potential both to overcome the limitations on enzyme availability and to design new biocatalysts that can improve bioprocesses and facilitate the realization of innovative biotransformations. The great challenge will be to incorporate these new processes to improve the biotechnological applications of archaeal enzymes and biomolecules.
ACKNOWLEDGEMENTS
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação Carlos Chagas Filho de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ).
Submitted: March 25, 2007; Returned to authors for corrections: July 16, 2007; Approved: July 29, 2007
- 1. Almeida, R.V.; Alqueres, S.M.C.; Larentis, A.L.; Rossle, S.C.S.; Cardoso, A.M.; Almeida, W.I.; Bisch, P.M.; Alves, T.L.M.; Martins, O.B. (2006). Cloning, expression, partial characterization and structural modeling of a novel esterase from Pyrococcus furiosus. Enz. Microb. Technol., 39, 1128-1136.
- 2. Amann, R.I.; Ludwig, W.; Schleifer, K.H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev., 59(1), 143-169.
- 3. Auchtung, T.A.; Takacs-Vesbach, C.D.; Cavanaugh, C.M. (2006). 16S rRNA phylogenetic investigation of the candidate division "Korarchaeota". Appl. Environ. Microbiol., 72(7), 5077-5082.
- 4. Bagai, R.; Madamwar, D. (1997). Continuous production of halophilic a-amylase through whole cell immobilization of Halobacterium salinarium. Appl. Biiochem. Biotechnol, 62(2), 213-218.
- 5. Beja, O.; Suzuki, M.T.; Koonin, E.V.; Aravind, L.; Hadd, A.; Nguyen, L.P.; Villacorta, R.; Amjadi, M.; Garrigues, C.; Jovanovich, S.B.; Feldman, R.A.; DeLong EF. (2000). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol., 2(5), 516-529.
- 6. Brown, J.R.; Doolittle, W.F. (1997). Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev., 61(4), 456-502.
- 7. Cardoso, A.M.; Clementino, M.B.M.; Martins, O.B.; Vieira, R.P.; Almeida, R.V.; Alquéres, S.M.C.; Almeida, W.I. (2003). Archaea: Potencial Biotecnológico. Rev. Biotec. Cien. Des., 30, 71-77.
- 8. Cardoso, A.M.; Polycarpo, C.; Martins, O.B.; Soll, D. (2006). A non-discriminating aspartyl-tRNA synthetase from Halobacterium salinarum. RNA Biology, 3, 110-114.
- 9. Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. (2002). Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol., 13(3), 253-261.
- 10. Chaban, B.; Ng, S.Y.; Jarrel, F.K. (2006). Archaeal habitats from the extreme to the ordinary. Can. J. Microbiol., 52, 73-116.
- 11. Clementino, M.M.; Fernandes, C.C.; Vieira, R.P.; Cardoso, A.M.; Polycarpo, C.R.; Martins O.B. (2007). Archaeal diversity in naturally occurring and impacted environments from a tropical region. J. Appl. Microbiol, 103(1), 141-151.
- 12. Costerton, J.W.; Lewandowski, Z.; DeBeer, D.; Caldwell, D.; Korber, D.; James, G. (1994). Biofilms, the customized microniche. J. Bacteriol, 176, 2137-2142.
- 13. DeLong, E.F. (1992). Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA, 89, 5685-5689.
- 14. Dennis, P.P. (1997). Ancient ciphers: translation in Archaea. Cell., 89, 1007-1010.
- 15. Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. (2005). Diversity of the human intestinal microbial flora. Science, 308 (5728), 1635-1638.
- 16. Eilers, H.; Pernthaler, J.; Glockner, F.O.; Amann, R. (2000). Culturability and In situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol, 66, 3044-3051.
- 17. Hanzawa, S.; Hoaki, T.; Jannasch, H.W.; Maruyama, T. (1996). An extremely thermostable serine protease from a hyperthermophilic archaeon Desulfurococcus strain SY, isolated from a deep-sea hydrothermal vent. J. Mar. Biotechnol, 4, 121-126.
- 18. Holmes, M.L.; Scopes, R.K.; Moritz, R.L.; Simpson, R.J.; Englert, C.; Pfeifer, F.; Dyall-Smith, M.L. (1997). Purification and analysis of an extremely halophilic b-galactosidase from Haloferax alicantei. Biochim. Biophys. Acta, 1337(2), 276-286.
- 19. Huber, H.; Hohn, M.J.; Rachel, R.; Fuchs, T.; Wimmer, V.C.; Stetter, K.O. (2002). A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature, 417, 63-67.
- 20. Ikeda, M.; Clark, D.S. (1998). Molecular cloning of extremely thermostable esterase gene from hyperthermophilic archaeon Pyrococcus furiosus in Escherichia coli. Biotechnol. Bioeng., 57(5), 624-629.
- 21. Kim, J.; Dordick, J.S. (1997). Unusual salt and solvent dependence of a protease from an extreme halophile. Biotechnol. Bioeng, 55(3), 471-479.
- 22. Koyama, K.; Yamaguchi, N.; Miyasaka, T. (1994). Antibody-mediated bacteriorhodopsin orientation for molecular device architectures. Science, 265, 762-764.
- 23. Ladenstein, R.; Antranikian, G. (1998). Proteins from hyperthermophiles: stability and enzymatic catalysis close to the boiling point of water. Adv. Biochem. Eng. Biotechnol., 61, 37-85.
- 24. Mayr, J.; Lupas, A.; Kellermann, J.; Eckerskorn, C.; Baumeister, W.; Peters, J. (1996). A hyperthermostable protease of the subtilisin family bound to the surface of the layer of the archaeon Staphylothermus marinus. Curr. Biol., 6, 739-749.
- 25. Niehaus, F.; Bertoldo, C.; Kähler, M.; Antranikian, G. (1999). Extremophiles as a source of novel enzymes for industrial application. Appl. Microbiol. Biotechnol., 51, 711-729.
- 26. Niehaus, F.; Peters, A.; Groudieva, T.; Antranikian, G. (2000). Cloning, expression and biochemical characterisation of a unique thermostable pullulan-hydrolysing enzyme from the hyperthermophilic archaeon Thermococcus aggregans. FEMS Microbiol. Lett., 190(2), 223-229.
- 27. Olsen, G.J. (1994). Archaea, Archaea, Everywhere. Nature, 371, 657.
- 28. Olsen, G.J.; Woese, C.R. (1997). Archaeal genomics: an overview. Cell, 89(7), 991-994.
- 29. Oren, A. (1999). Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev., 63, 334-348.
- 30. Ovreas, L.; Forney, L.; Daae, F.L.; Torsvik, V. (1997). Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol, 63, 3367-3373.
- 31. Patel, G.B.; Zhou, H.; KuoLee, R.; Chen, W. (2004). Archaeosomes as adjuvants for combination vaccines. J. Liposome Res., 14(3), 191-202.
- 32. Perler, F.B.; Kumar, S.; Kong, H. (1996). Thermostable DNA polymerases. Adv. Protein Chem., 48, 377-435.
- 33. Polycarpo, C.; Sheppard, K.; Randau, L.; Ambrogelly, A.; Cardoso, A.M.; Fukai, S.; Herring, S.; Hohn, M.J.; Nakamura, Y.; Oshikane, H.; Palioura, S.; Salazar, J.C.; Yuan, J.; Nureki, O.; Soll, D. (2007). Features of Aminoacyl-tRNA Synthesis Unique to Archaea. In: Cavicchioli, R. (ed). Archaea: Molecular and Cellular Biology. Washington, D.C. - USA, p.198-208.
- 34. Pondexter, J.S.; Leadbetter, E.R. (1986). Enrichment cultures in microbial ecology. In: Poindexter, J.S.; Leadbetter, E.R. (eds). Bacteria in Nature Plenum Press, New York, USA, p.229-260.
- 35. Robertson, C.E.; Harris, J.K.; Spear, J.R.; Pace, N.R. (2005). Phylogenetic diversity and ecology of environmental Archaea. Curr Opin Microbiol., 8, 638-642.
- 36. Rondon, M.R.; August, P.R.; Bettermann, A.D.; Brady, S.F.; Grossman, T.H.; Liles, M.R.; Loiacono, K.A.; Lynch, B.A.; MacNeil, I.A.; Minor, C.; Tiong, C.L.; Gilman, M.; Osburne, M.S.; Clardy, J.; Handelsman, J.; Goodman, R.M. (2000). Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol, 66(6), 2541-2547.
- 37. Russell, N.J. (2000). Toward a molecular understanding of cold activity of enzymes from psychrophiles. Extrem., 4(2), 83-90.
- 38. Ryu, K.; Kim, J.; Dordick, J.S. (1994). Catalytic properties and potential of an extracellular protease from an extreme halophile. Enzyme Microb. Technol., 16(4), 266-275.
- 39. Sandaa, R.A.; Enger, O.; Torsvik, V. (1999). Abundance and diversity of Archaea in heavy-metal-contaminated soils. Appl. Environ. Microbiol, 65, 3293-3297.
- 40. Schiraldi, C.; Giuliano M.; de Rosa M. (2001). Perspectives on biotechnological applications of archaea. Archaea, 1, 75-86.
- 41. Schumacher, K.; Heine, E.; H. Hocker. 2001. Extremozymes for improving wool properties. J. Biotechnol., 89: 281-288.
- 42. Skillman, L.C.; Evans, P.N.; Strompl, C.; Joblin, K.N. (2006). 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett. Appl. Microbiol., 42(3), 222-228.
- 43. Sleytr, U.B.; Sara, M. (1997). Bacterial and archaeal S-layer proteins: structure-function relationships and their biotechnological applications. Trends Biotechnol., 15(1), 20-26.
- 44. Southworth, M.W.; Kong, H.; Kucera, R.B.; Ware, J.; Jannasch, H.W.; Perler, F.B. (1996) Cloning of thermostable DNA polymerases from hyperthermophilic marine Archaea with emphasis on Thermococcus sp 9 degrees N-7 and mutations affecting 3'-5'- exonuclease activity. Proc Natl Acad Sci USA, 93, 5281-5285.
- 45. Sprott, G.D.; Tolson, D.L.; Patel, G.B. (1997). Archaeosomes as novel antigen delivery systems. FEMS Microbiol Lett., 154(1), 17-22.
- 46. Synowiecki, J.; Grzybowska, B.; Zdzieblo, A. (2006). Sources, properties and suitability of new thermostable enzymes in food processing. Crit. Rev. Food. Sci. Nutr., 46(3), 197-205.
- 47. Vieira, R.P.; Clementino, M.M.; Cardoso, A.M.; Oliveira, D.N.; Albano, R.M.; Gonzalez, A.M.; Paranhos, R.; Martins, O.B. (2007). Archaeal Communities in a Tropical Estuarine Ecosystem: Guanabara Bay, Brazil. Microb Ecol, DOI: 10.1007/s00248-007-9261-y
- 48. Voëlkl, P.; Markiewicz, P.; Stetter, K.O.; Miller, J.H. (1995). The sequence of a subtilisin-type protease (aerolysin) from the hyperthermophilic archaeon Pyrobaculum aerophilum reveals sites important to thermostability. Protein Sci., 3, 1329-1340.
- 49. Voorhorst, W.G.B.; Eggen, R.I.L.; Geerling, A.C.M.; Platteeuw, C.; Siezen, R.J.; Vos, W.M. (1996). Isolation and characterization of the hyperthermostable serine protease, pyrolysin, and its gene from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem, 271, 20426-20431.
- 50. Woese, C.R. (1998). The universal ancestror. Proc. Natl. Acad. Sci. USA, 95, 6854-6859.
- 51. Woese, C.R.; Fox, G.E. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA, 74, 5088-5090.
- 52. Yuan, J.; Palioura, S.; Salazar, J.C.; Su, D.; ODonoghue, P.; Hohn, M.J.; Cardoso, A.M.; Whitman, W.B.; Soll, D. (2006). RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc Natl Acad Sci USA, 103, 18923-1892.
Publication Dates
-
Publication in this collection
17 Oct 2007 -
Date of issue
Sept 2007
History
-
Accepted
29 July 2007 -
Reviewed
16 July 2007 -
Received
25 Mar 2007