Abstracts
In order to study the epidemiology of Salmonella Enteritidis outbreaks and determine the source of contamination so that a recurrence can be avoided, detailed characterization is necessary. Thus, the purpose of this study was to verify whether rep-PCR was able to discriminate among Salmonella Enteritidis isolates. Phage typing, detection of virulence genes and antimicrobial resistance testing were also associated to rep-PCR results. One hundred and two S. Enteritidis isolates from broiler carcasses, food, human, pigs, poultry-related samples, and nine isolates from other countries were genotypically typed by REP-PCR, ERIC-PCR and BOX-PCR, collectively called rep-PCR. Phage typing, detection of virulence genes and antimicrobial resistance testing were also performed. Only three fingerprinting profiles were obtained with each rep-PCR method, with the majority of isolates belonging to the same profile. No relationship was observed between genotypic profile and year, place of isolation or source of infection. However, the less frequent rep-PCR profiles showed single antimicrobial resistance patterns. Although few strains isolated from swine were analyzed, different antimicrobial resistance patterns were observed. Furthermore, phage type 4 was not found in swine isolates. rep-PCR showed a lower discriminatory power as compared with antimicrobial resistance and phage typing, but the combination of genotypic and phenotypic methods was more discriminatory than any method alone, resulting in 48 different types.
Salmonella Enteritidis; rep-PCR; phage typing; antimicrobial resistance; virulence genes
Uma caracterização detalhada de Salmonella Enteritidis é necessária para que possa ser desenvolvido o estudo da epidemiologia dos surtos causados por este organismo, bem como a determinação da fonte de contaminação, evitando que ocorram novos surtos. Assim, o objetivo deste estudo foi verificar se a rep-PCR era capaz de diferenciar isolados de S. Enteritidis. A fagotipagem, a detecção de genes de virulência e a determinação de resistência antimicrobiana foram associadas aos resultados da rep-PCR. Cento e duas S. Enteritidis isoladas de carcaças de frango, alimentos prontos para consumo, humanos suínos, amostras relacionadas a aves, e nove isolados de outros países foram genotipicamente tipados por REP-PCR, ERIC-PCR e BOX-PCR, juntamente chamados de rep-PCR. A fagotipagem, a detecção de genes de virulência e a determinação de resistência antimicrobiana também foram realizadas. Somente três padrões de fingerprinting foram obtidos com cada método de rep-PCR, sendo que a maioria dos isolados pertenceu ao mesmo perfil. Nenhuma relação foi observada entre o perfil genotípico e o ano, o local de isolamento e a fonte de infecção. Entretanto, os perfis menos freqüentes de rep-PCR apresentaram padrões de resistência antimicrobiana únicos. Embora poucas amostras de suínos tenham sido analisadas, diferentes padrões de resistência antimicrobiana foram observados. Além disso, o fagotipo 4 não foi encontrado em isolados de suínos. A rep-PCR apresentou um menor poder discriminatório quando comparada com a resistência antimicrobiana e com a fagotipagem, mas a combinação dos métodos genotípicos e fenotípicos foi mais discriminatória do que qualquer método isolado, resultando em 48 tipos diferentes.
Salmonella Enteritidis; rep-PCR; fagotipagem; resistência antimicrobiana e genes de virulência
GENERAL MICROBIOLOGY
Phenotypic and genotypic characterization of Salmonella Enteritidis isolates
Caracterização fenotípica e genotípica de isolados de Salmonella Enteritidis
Sílvia Dias de OliveiraI,II* * Corresponding Author. Mailing address: Faculdade de Biociências, Pontifícia Universidade Católica do Rio Grande do Sul (PUCRS), Av. Ipiranga 6681, 90619-900, Porto Alegre, Brasil. Tel.: +55-51-30263242; Fax: +55-51-33087305. E-mail: silviadias@pucrs.br ; Marjo Cadó BessaIV; Luciana Ruschel dos SantosV; Marisa Ribeiro de Itapema CardosoIV; Adriano BrandelliII; Cláudio Wageck CanalIII
IFaculdade de Biociências, Pontifícia Universidade Católica do Rio Grande do Sul, Porto Alegre, RS, Brasil
IIDepartamento de Ciência de Alimentos, Instituto de Ciência e Tecnologia de Alimentos, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brasil
IIISetor de Virologia, Faculdade de Veterinária, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brasil
IVSetor de Medicina Veterinária Preventiva, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brasil
VUniversidade de Passo Fundo, Passo Fundo, RS, Brasil
ABSTRACT
In order to study the epidemiology of Salmonella Enteritidis outbreaks and determine the source of contamination so that a recurrence can be avoided, detailed characterization is necessary. Thus, the purpose of this study was to verify whether rep-PCR was able to discriminate among Salmonella Enteritidis isolates. Phage typing, detection of virulence genes and antimicrobial resistance testing were also associated to rep-PCR results. One hundred and two S. Enteritidis isolates from broiler carcasses, food, human, pigs, poultry-related samples, and nine isolates from other countries were genotypically typed by REP-PCR, ERIC-PCR and BOX-PCR, collectively called rep-PCR. Phage typing, detection of virulence genes and antimicrobial resistance testing were also performed. Only three fingerprinting profiles were obtained with each rep-PCR method, with the majority of isolates belonging to the same profile. No relationship was observed between genotypic profile and year, place of isolation or source of infection. However, the less frequent rep-PCR profiles showed single antimicrobial resistance patterns. Although few strains isolated from swine were analyzed, different antimicrobial resistance patterns were observed. Furthermore, phage type 4 was not found in swine isolates. rep-PCR showed a lower discriminatory power as compared with antimicrobial resistance and phage typing, but the combination of genotypic and phenotypic methods was more discriminatory than any method alone, resulting in 48 different types.
Key-words: Salmonella Enteritidis, rep-PCR, phage typing, antimicrobial resistance, virulence genes
RESUMO
Uma caracterização detalhada de Salmonella Enteritidis é necessária para que possa ser desenvolvido o estudo da epidemiologia dos surtos causados por este organismo, bem como a determinação da fonte de contaminação, evitando que ocorram novos surtos. Assim, o objetivo deste estudo foi verificar se a rep-PCR era capaz de diferenciar isolados de S. Enteritidis. A fagotipagem, a detecção de genes de virulência e a determinação de resistência antimicrobiana foram associadas aos resultados da rep-PCR. Cento e duas S. Enteritidis isoladas de carcaças de frango, alimentos prontos para consumo, humanos suínos, amostras relacionadas a aves, e nove isolados de outros países foram genotipicamente tipados por REP-PCR, ERIC-PCR e BOX-PCR, juntamente chamados de rep-PCR. A fagotipagem, a detecção de genes de virulência e a determinação de resistência antimicrobiana também foram realizadas. Somente três padrões de fingerprinting foram obtidos com cada método de rep-PCR, sendo que a maioria dos isolados pertenceu ao mesmo perfil. Nenhuma relação foi observada entre o perfil genotípico e o ano, o local de isolamento e a fonte de infecção. Entretanto, os perfis menos freqüentes de rep-PCR apresentaram padrões de resistência antimicrobiana únicos. Embora poucas amostras de suínos tenham sido analisadas, diferentes padrões de resistência antimicrobiana foram observados. Além disso, o fagotipo 4 não foi encontrado em isolados de suínos. A rep-PCR apresentou um menor poder discriminatório quando comparada com a resistência antimicrobiana e com a fagotipagem, mas a combinação dos métodos genotípicos e fenotípicos foi mais discriminatória do que qualquer método isolado, resultando em 48 tipos diferentes.
Palavras-chave: Salmonella Enteritidis, rep-PCR, fagotipagem, resistência antimicrobiana e genes de virulência
INTRODUCTION
There has been a dramatic increase in the incidence of Salmonella Enteritidis worldwide since 1980 (3,14,25,28). In Brazil, the increase in the incidence of S. Enteritidis from foodborne outbreaks, human infections, non-human sources, broiler carcasses and other poultry materials has been reported since the 90's (5,6,25,29,33,34), and poultry is likely the main source of human infections (10). Typing of bacteria can be used to determine whether isolates recovered from different patients or from the environment are related and, in so doing, provide evidence for a common source of transmission of the agent (9). Many phenotypic and genotypic typing methods have been applied to epidemiologically trace S. Enteritidis infections. Traditional epidemiological methods include biotyping, serotyping and phage typing of isolates, as well as antimicrobial resistance testing, although these methods do not always give enough information for epidemiological purposes. The limitations of phenotypic analysis have led to the progressive development of genotypic strategies.
Genotypic methods include analysis of plasmid content and plasmid restriction patterns (17), random amplified polymorphic DNA analysis (14), IS 200 typing and ribotyping (20), restriction fragment length polymorphism (RFLP) (36), pulsed-field gel electrophoresis (PFGE) (7,15,35), amplified-fragment length polymorphism (AFLP) (15), multilocus sequence typing of bacterial DNA (19) and repetitive-sequence polymerase chain reaction (rep-PCR) (26,38,39).
Among the molecular methods for typing bacteria, the REP-, ERIC- and BOX-PCR (collectively referred to as rep-PCR) genomic fingerprinting have been found to be extremely reliable, reproducible, rapid and highly discriminatory (38,39). This method is based on the amplification of DNA sequences by PCR with primer sets complementary to naturally occurring, highly conserved, repetitive DNA sequences present in multiple copies in distinct intergenic positions in the genomes of most Gram negative and several Gram positive bacteria (18).
The purpose of the present study was to asses the level of genetic diversity and possible relationships among S. Enteritidis strains isolated from food, human, pigs, broiler carcasses and poultry-related samples using rep-PCR. Phage typing, detection of virulence genes and antimicrobial resistance testing were associated with the results of rep-PCR.
MATERIALS AND METHODS
Bacterial isolates and growth conditions
Our study was carried out using a total of 111 Salmonella Enteritidis isolates, being 102 isolates from Southern Brazil (Rio Grande do Sul and Santa Catarina States) collected over the period 1995-2001 (Table 1). Seventeen isolates were from human samples; 31 from food involved in foodborne outbreaks; 22 from broiler carcasses; 11 from pigs (lymph nodes, faeces and fresh pork sausage); and 21 were from other poultry-related samples (viscera and environmental samples). Human and food isolates did not belong to the same outbreak. Furthermore, nine epidemiologically unrelated isolates from other countries (Zimbabwe, Egypt, Tanzania, Italy and Albania) were analyzed. An outgroup of four different serovars of Salmonella (Panama, Bredeney, Agona and Typhimurium) and S. Enteritidis ATCC 13076 were also included. The isolates from human samples, food, broiler carcasses and poultry had been phage typed in a previous study (31) and had also their antimicrobial resistance determined (23). For genotyping analysis, all isolates were cultured in TSB and incubated overnight at 37ºC.
Phage typing
Seven of 11 isolates from swine and the isolates from other countries were phage typed in this study. Phage typing was made as described by Santos et al. (2003).
Antimicrobial susceptibility testing
The 11 S. Enteritidis isolated from swine and the nine isolates from other countries were tested for antimicrobial susceptibility. The test was performed according to the guidelines of the National Committee for Clinical Laboratory Standards (22) for the disk diffusion technique using commercial disks. The antimicrobials tested were: ampicillin (Ampc), cephalotin (Cef), ciprofloxacin (Cipx), chloramphenicol (Chl), gentamicin (Gen), streptomycin (Str), nitrofurantoin (Nit), norfloxacin (Nor), nalidixic acid (Nalx), tetracycline (Tet), and sulphonamide (Smx). Inhibition zones were measured and scored as sensitive, intermediate and resistant according to the NCCLS recommendations. Escherichia coli ATCC 25922 was used as a reference strain for antibiotic disc control.
Detection of virulence genes
The detection of invA, spvR and spvC virulence genes was performed in the isolates from other countries. These genes were detected by PCR as previously described for other S. Enteritidis strains that belonged to this study (24).
rep-PCR fingerprinting
A 1 mL aliquot of culture was centrifuged at 8,000 g for 10 min. Cells were washed twice in 1 mL of 1 M NaCl and pelleted by centrifugation at 8,000 g for 10 min. Cells were resuspended in 100 mL of TE (10 mM Tris HCl pH 8.0, 1 mM EDTA). Total DNA was prepared according to the method of Rademaker and de Bruijn (1997). Briefly, bacterial cells were lysed with 500 mL of 5 M guanidine thiocyanate, 0.03 M N-lauroyl sarkosine, 0.1 M EDTA, for 5 min at 4ºC. After, 250 mL cold 7.5 M ammonium acetate were added, tubes were gently shaken and incubated for 5 min at 4ºC. An aliquot of 500 mL of chloroform/iso-amyl-alcohol (24:1) was added and the mixture was vortexed vigorously. After centrifugation at 16,000 g for 10 min, the DNA-containing pellet was further washed with isopropyl alcohol. DNA concentration was determined by spectrofluorimetry at excitation and emission wavelenghts of 365 nm and 460 nm, respectively, using the DNA-specific dye Hoechst 33258 (Amersham Pharmacia Biotech), in a DyNA QuantTM 200 fluorometer (Hoefer Pharmacia Biotech Inc.) according to manufacturer's instructions. DNA concentration was adjusted to 200 ng/mL. REP (REP1R-I and REP2-I), ERIC (ERIC1R and ERIC2) and BOX (BOXA1R) primer sequences (Life Technologies) were as previously described (39). PCR was performed in a 25 mL reaction volume containing Gitschier buffer (16.6 mM (NH4)2SO4, 67 mM Tris-HCl pH 8.8, 6.7 mM MgCl2, 6.7 mM EDTA pH 8.8, 30 mM b-mercapto-ethanol), 1.25 mM of each deoxynucleotide (GibcoBRL), 2 U of Taq DNA Polymerase (Cenbiot Enzimas) and 200 ng of template DNA. Fifty rmol/mL, 70 rmol/mL and 100 rmol/mL of primers for REP, ERIC and BOX-PCR, respectively were added. Amplifications were carried out using a GeneAmp PCR System 2400 Thermocycler (Perkin Elmer Instruments). For REP-PCR primers, cycles were as follows: initial denaturation at 95ºC for 5 min, followed by 35 cycles of denaturation at 94ºC for 1 min, annealing at 40ºC for 1 min and extension at 65ºC for 8 min, with a final extension at 65ºC for 16 min. For ERIC-PCR primers, the cycles were the same except for the 50ºC annealing temperature. However, for BOX-PCR primers, cycles were as follows: initial denaturation at 97ºC for 5 min, followed by 30 cycles of denaturation at 94ºC for 1 min, annealing at 53ºC for 1 min and extension at 65ºC for 8 min, with a final extension at 65ºC for 16 min. Negative control reactions that contained every component except the target DNA were included in each experimental set. The S. Enteritidis ATCC 13076 was included in each set of reactions as parameter interreactions. Amplification products were electrophoresed on 1.5% agarose gels at 5 V/cm. Gels were stained with ethidium bromide (0.5 mg/mL) and visualized on a UV transiluminator. Sizes of amplicons were determined by comparison with a concurrently run DNA molecular size marker (100-bp DNA ladder, Invitrogen). Each experiment was repeated at least twice.
Analysis of amplicon patterns
Gel images containing the profiles were captured by the imaging system Ultra-Lum (Paramount, CA). The Quantity One version 4.5 software (Bio-Rad) was used for band detection, normalization, and matching. A 5% tolerance level for matching was allowed. The REP, ERIC and BOX-PCR fingerprinting were transformed into binary code depending on the presence or absence of each band. Similarity between profiles was determined using the Jaccard coefficient and dendrograms were generated by the unweighted pair-group with mathematic average (UPGMA) method. All calculations were performed using a NTSYS-PC version 1.8 software (Applied Biostatistics).
Discriminatory power of the methods
The discriminatory power was measured by the Simpson's index of diversity (D) that indicates the average probability that a typing system will assign a different type to two unrelated strains randomly sampled from a population (8).
RESULTS
Phage typing, antimicrobial resistance and detection of virulence genes
The phage typing data are summarized in Table 1. Overall, eight different phage types were detected. In the S. Enteritidis isolates previously analyzed, four phage types (PT4, PT4a, PT6a and PT7) had been identified (31). In this study, four additional phage types were detected (PT7a, PT6, PT11 and PT9), being three different phage types detected in the seven swine S. Enteritidis isolates from the same slaughterhouse, and none was PT4. The Simpson's index of diversity (D) for phage typing method was 0.6. Twelve antimicrobial resistance patterns were found in isolates from pig and other countries (Table 1). Resistance to cephalotin, ciprofloxacin, gentamicin, and norfloxacin was not detected. Considering all 111 isolates, 27 different resistance patterns were observed, showing a D value of 0.86. The virulence genes invA, spvR and spvC were detected in all isolates from other countries. Taking together all 111 isolates, the virulence genes invA, spvR and spvC were detected in 100%, 91.2% and 91%, respectively.
rep-PCR fingerprinting
Profiles could be discriminated by the number and position of amplified DNA bands. In order to determine the reproducibility of PCR fingerprinting, duplicate samples were analyzed at different times. Electrophoreses in agarose gels were performed at the same time, with each isolate running side-by-side. Different patterns were not obtained from the same sample, although differences in band intensity were sometimes observed. The very weak bands were not reproducible, and were therefore not taken into account. Many bands were larger than about 1500 bp and difficult to resolve. Bands between about 140 and 1500 bp were less numerous, better resolved, and therefore, useful for matching fingerprints. Most rep-PCR profiles were similar, except for the presence or absence of one or few DNA fragments.
REP-PCR fingerprinting
With the primer combination REP1R4-I and REP2-I, three different profiles were found: R1, R2 and R3 (Fig. 1). The number of bands per profile varied from 9 to 12 bands ranging between about 140 bp to 1340 bp. Conserved fragments of approximately 235, 590, 725, 785, 985, 1050 and 1340 bp were found in all S. Enteritidis isolates. Weak fragments of approximately 450 and 625 bp were also found in the same isolates. The fragments of approximately 235, 590, 725, 785, 985 and 1050 were also conserved in the other serovars tested. Profile R1 showed all bands mentioned above. Profiles R2 and R3 showed the same bands as R1, with some additional bands. Profile R2 was characterized by the presence of one weak fragment of approximately 135 bp and two additional fragments of approximately 305 and 500 bp. Profile R3 showed two additional fragments as compared to R1 of approximately 160 and 350 bp. The three profiles were clustered at 71.4% similarity. Profiles R1 and R3 formed a single cluster at 81.8% similarity. The S. Typhimurium isolate displayed profile R1, but the other serovars formed a different cluster with similarity below 70%. Most of the isolates (108/111) belonged to profile R1, while profile R2 was found only in one of the isolates from Tanzania (isolate 111) and R3 in two isolates from broiler carcasses (isolates 14 and 15). The D value for this typing method was 0.05.
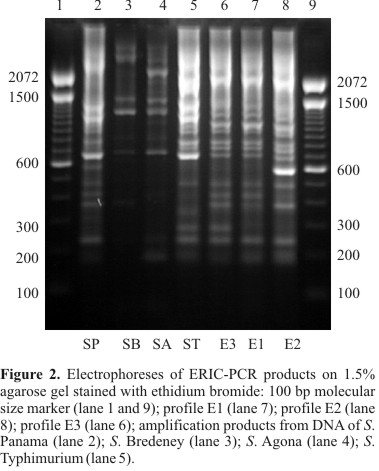
ERIC-PCR fingerprinting
With the ERIC set of primers, three profiles were also observed and referred to as E1, E2 and E3 (Fig. 2). The size of amplification products ranged from about 190 bp to 1430 bp forming profiles with 12 or 13 bands each. Profile E1 showed fragments of approximately 190, 235, 370, 415, 480, 685, 730, 800, 925, 1040, 1150 and 1390 bp. These fragments were also found in profiles E2 and E3. Conserved amplicons of approximately 235, 685 and 800 bp were also found in the other serovars tested. Profile E2 differed from E1 by the presence of one intense fragment of approximately 590 bp. Profile E3 also showed only one additional fragment of approximately 275 bp. Whereas profile E1 was found in 110 S. Enteritidis isolates, only one isolate from food (isolate 59) belonged to profile E2, and profile E3 was only present in the ATCC strain. Profiles E1, E2 and E3 formed a single cluster at 89% similarity level, being E1 and E2 clustered at 92.3% similarity. S. Typhimurium and S. Panama isolates showed similarity with the cluster formed by E1, E2 and E3 at 79.2% and 75%, respectively. S. Agona and S. Bredeney isolates formed a different cluster at similarity below 70%. The D value for this typing method was 0.04.
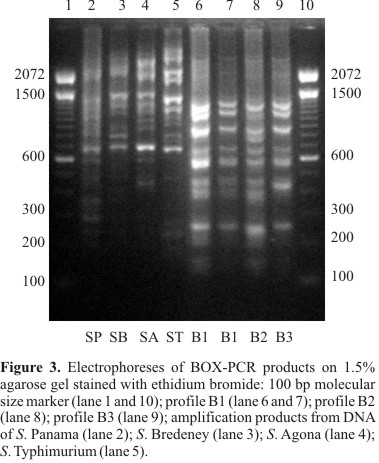
BOX-PCR fingerprinting
By BOX-PCR with primer BOXA1R, three different profiles were found: B1, B2 and B3 (Fig. 3). The size of amplification products ranged from about 140 bp to 1230 bp. Profiles showed eight or nine bands. All S. Enteritidis isolates were characterized by the presence of amplicons of approximately 140, 245, 440, 565, 665, 860 and 1230 bp. A conserved fragment of approximately 665 bp was also found in the other serovars. Profile B1 showed nine DNA bands as follows: approximately 140, 245, 380, 440, 565, 665, 860, 1080 and 1230 bp. Profile B2 differed from B1 by the presence of one fragment of approximately 750 bp and the absence of the fragment of approximately 1080 bp. The profile B3 showed the same fragments as B1, except for fragment of 380 bp. The majority of the S. Enteritidis isolates belonged to profile B1. Profile B2 was represented only by one isolate from swine (isolate 99). BOX-PCR, as well as REP-PCR, discriminated one of the isolates from Tanzania (isolate 111), which formed alone the profile B3. All S. Enteritidis were clustered at 85.6% similarity. Profiles B1 and B3 formed a single cluster with 94.1% similarity. The S. Enteritidis and other serovars were clustered at below 70% of similarity. The D value for this typing method was 0.04.
Combination of the typing methods
The combination of the results described above with those obtained in previous studies for phage typing, presence of virulence genes and antimicrobial resistance pattern (23,24,31) permitted the identification of a total of 48 types (Table 1). Types 1, 2, 7, 8 and 9 were the most prevalent, being found in 56 isolates. No type was found in all the different sources (broiler carcasses, food, human samples, poultry and swine samples). Types 2 and 7 were more disseminated between sources, being the former found in broiler carcasses, food, human samples and poultry, and the latter found in broiler carcasses, food, human samples and isolates from other countries. Types 1, 2, 7, 8, 9 and 18 were the only types found in more than one source. The data obtained afforded the construction of a dendrogram to show similarity levels between isolates (Fig. 4).
DISCUSSION
It is of great public health significance that strains of S. Enteritidis can be rapidly identified and distinguished; therefore in the present study we investigated the suitability of rep-PCR fingerprinting to discriminate among S. Enteritidis isolates. Furthermore, we assessed whether the combination of rep-PCR with phage typing, presence of virulence genes and antimicrobial resistance could increase the discrimination between the isolates.
PCR fingerprinting with three primer sets showed that the majority of the isolates belonged to the same profile (R1E1B1). Most of these isolates had been formerly analyzed by Santos (2001), who obtained only one pattern using RAPD with primers OPB17 and 1254. These results can be explained in three different ways: the majority of isolates belong to the same clonal lineage, rep-PCR cannot discriminate among them, or both hypothesis. Probably, the third hypothesis is the most probable, since the majority of the S. Enteritidis isolates analyzed belonged to PT4 or PT4a, considered highly clonal. Therefore, the high homogeneity detected among isolates, even with those obtained from other countries and thus unrelated with the local strains, indicates the low discriminatory power of rep-PCR for S. Enteritidis.
The great heterogeneity found by Chmielewski et al. (2002) using REP- and ERIC-PCR in 31 S. Enteritidis isolates stimulated our study. According to these authors, the two methods were shown to be highly discriminatory and useful for epidemiological evaluation of the serovar Enteritidis. The differences found in REP-PCR can be due to the fact that the REP primers used were different from those used in the present study. However, the ERIC primers were the same and the protocol for DNA purification was similar, which does not justify the differences found in the ERIC-PCR results from the two works. Weigel et al. (2001) also used rep-PCR with the same primers tested in the present study to type Salmonella and compared the profiles obtained with those generated by PFGE after DNA cleavage with three different enzymes. According to these authors, both methods were equivalent in detecting genetic variance and neither revealed improbable patterns of transmission. However, these authors did not specify which Salmonella serovars were tested, and probably they analyzed serovars with more genetic variance as compared to Enteritidis. A high discriminatory power also was found by Rasschaert et al. (2005), since divided 13 S. Enteritidis into six clusters using the same ERIC primer set used in the present study. However, the differences found in the fingerprints of these strains consisted in bands of different intensity at 2000 bp, a size that was not analyzed in the present study, because bands larger than about 1500 bp displayed low resolution power in our experiments, what can explain the differences from our results.
The high level of similarity between S. Enteritidis and S. Typhimurium detected by REP-PCR is in accordance with findings of Millemann et al. (1996) for ERIC-PCR. They identified just two patterns that differed by a single band in 56 S. Typhimurium and 14 S. Enteritidis isolated from poultry. All 14 S. Enteritidis and seven S. Typhimurium isolates generated the same pattern. Our results and the findings of Millemann et al. (1996) and Burr et al. (1998), using the same primers, are in disagreement with those described by Van Lith and Aarts (1994) and Koh-Luar et al. (1998), who found each serotype to be characterized by a unique DNA profile. Burr et al. (1998) characterized other Salmonella serovars and found that every isolate had a unique fingerprinting but the serovars were not grouped together in major branches. Thus, serotypes were not identified by ERIC-PCR.
The low discriminatory power of rep-PCR for typing S. Enteritidis described in the present study was also found by other authors that used other genotypic methods as PFGE, considered the gold standard for Salmonella typing (7,12,13). Despite, one Brazilian study using ribotyping discriminated 14 ribotypes, six being identified within PT 8 and three within PT 4 isolates (4).
The high prevalence of virulence genes together with the genetic homogeneity between S. Enteritidis isolates detected in the present study are in accordance with Burr et al. (1998) that suggested that the requirement for invasion and virulence factors selected a more homogeneous subpopulation.
No correlation between fingerprints, year and place of isolation could be traced. We were also unable to correlate a particular rep-PCR profile with PT, with exception of the PT9 isolate, which also showed unique profiles in REP and BOX-PCR. However, there was a correlation between the less frequent rep-PCR profiles and unique antimicrobial resistance patterns (Table 1). Although few strains isolated from swine were analyzed, we observed differences in the antimicrobial resistance pattern of this isolates that are probably a consequence of the different drugs used in chicken and swine rearing. Furthermore, phage type 4 was not found in swine. rep-PCR showed a lower discriminatory power as compared to phage typing and antimicrobial resistance. However, the combination of the different methods improved the discriminatory power among S. Enteritidis isolates, even though the same antimicrobial resistance pattern did not necessarily reflect genetic relatedness. The collection of 111 S. Enteritidis isolates was further differentiated into 48 types by combining the rep-PCR patterns with phage types, presence of virulence genes and antimicrobial resistance profiles. When all methods were combined, the majority of isolates from swine displayed single profiles, indicating that the source of the bacteria probably is not the same.
In conclusion, we found that rep-PCR performed as described in the present study was not useful for S. Enteritidis typing, because it was not able to discriminate potentially different isolates. However, the combined use of genotypic and phenotypic methods allowed a more accurate discrimination between isolates.
ACKNOWLEDGEMENTS
We thank S. Rubino from Universitá di Sassari for providing Salmonella Enteritidis from other countries. Financial support was provided by Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Submitted: February 28, 2007; Returned to authors for corrections: May 19, 2007; Approved: September 20, 2007.
- 1. Burr, M.D.; Josephson, K.L.; Pepper, I.L. (1998). An evaluation of ERIC PCR and AP PCR fingerprinting for discriminating Salmonella serotypes. Lett Appl. Microbiol., 27, 24-30.
- 2. Chmielewski, R.; Wieliczko, A.; Kuczkowski, M.; Mazurkiewicz, M.; Ugorski, M. (2002). Comparison of ITS profiling, REP- and ERIC-PCR of Salmonella Enteritidis isolates from Poland. J. Vet. Med, 49, 163-168.
- 3. Fantasia, M.; Filetici, E. (1994). Salmonella enteritidis in Italy. Int. J. Food Microbiol., 21, 7-13.
- 4. Fernandes S.A. et al (2003). Phenotypic and molecular characterization of Salmonella Enteritidis strains isolated in Sao Paulo, Brazil. Rev. Inst. Med. Trop. São Paulo; 45: 59-63.
- 5. Fuzihara, T.O.; Fernandes, S.A.; Franco, B.D. (2000). Prevalence and dissemination of Salmonella serotypes along the slaughtering process in Brazilian small poultry slaughter-houses. J. Food Prot., 63, 1749-1753.
- 6. Hofer, E.; Silva Filho, S.J.; Reis, E.M.F. (1997). Prevalência de sorovares de Salmonella isolados de aves no Brasil. Pesqui. Vet. Bras 17, 55-62.
- 7. Hudson, C.R.; Garcia, M.; Gast, R.K.; Maurer, J.J. (2001). Determination of close genetic relatedness of the major Salmonella enteritidis phage types by pulsed-field gel electrophresis and DNA sequence analysis of several Salmonella virulence genes. Avian Dis, 48, 875-886.
- 8. Hunter, P.R. (1990). Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol, 28, 1903-1905.
- 9. Kang, H.P.; Dunne, W.M. (2003). Stability of repetitive-sequence PCR patterns with respect to culture age and subculture frequency. J. Clin. Microbiol, 41, 2694-2696.
- 10. Khakhria, R.; Woodward, D.; Johnson, W.M.; Poppe, C. (1997). Salmonella isolated from humans, animals and other sources in Canada. Epidemiol. Infect, 119, 15-23.
- 11. Koh-Luar, S.I.; Choo, G.H.B.; Tan, S.L.R.; Tan, C.W.K. (1998). Genomic fingerprintings of Salmonella species generated with repetitive element sequence-based PCR. World J. Microbiol. Biotechnol, 14, 17-22.
- 12. Kotetishvili, M.; Stine, O.C.; Kreger, A.; Morris Jr., J.G.; Sulakvelidze, A. (2002). Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol, 40, 1626-1635.
- 13. Liebana, E.; Garcia-Migura, L.; Breslin, M.F.; Davies, R.H.; Woodward, M.J. (2001). Diversity of strains of Salmonella enterica serotype Enteritidis from English poultry farms assessed by multiple genetic fingerprinting. J. Clin. Microbiol., 39, 154-161.
- 14. Lin, A.W.; Usera, M.A.; Barret, T.J.; Goldsby, R.A. (1996). Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella enteritidis J. Clin. Microbiol, 34, 870-876.
- 15. Lindstedt, B.A.; Heir, E.; Vardund, T.; Kapperud, G. (2000). Fluorescent amplified-fragment length polymorphism genotyping of Salmonella enterica subsp. enterica serovars and comparison with pulsed-field gel electrophoresis typing. J. Clin. Microbiol, 38, 1623-1627.
- 16. Ling, J.M.; Koo, I.C.; Kam, K.M.; Cheng, A.F. (1998). Antimicrobial susceptibilities and molecular epidemiology of Salmonella enterica serotype Enteritidis strains isolated in Hong Kong from 1986 to 1996. J. Clin. Microbiol, 36, 1693-1699.
- 17. Liu, P.Y.; Lau, Y.J.; Hu, B.S.; Shyr, J.M.; Shi, Z.Y.; Tsai, W.S.; Lin, Y.H.; Tseng, C.Y. (1995). Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J. Clin. Microbiol 33, 1779-1783.
- 18. Lupski, J.R.; Weinstock, G.M. (1992). Short, interspersed repetitive DNA sequences in prokaryotic genomes. J. Bacteriol, 174, 4525-4529.
- 19. Maiden, M.C.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russel, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; Feavers, I.M.; Achtman, M.; Spratt, B.G. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A., 95, 3140-3145.
- 20. Millemann, Y.; Lesage-Descauses, M.C.; Chaslus-Dancla, E.; Lafont, J.P. (1995). Value of plasmid profiling, ribotyping and detection of IS200 for tracing avian isolates of Salmonella typhimurium and enteritidis J. Clin. Microbiol, 33, 173-179.
- 21. Millemann, Y.; Lesage-Descauses, M.C.; Lafont, J.P.; Chaslus-Dancla, E. (1996). Comparison of random amplified polymorphic DNA analysis and enterobacterial repetitive intergenic consensus-PCR for epidemiological studies of Salmonella FEMS Immunol. Med. Microbiol, 14, 129-134.
-
22NCCLS (2001). Performance standards for antimicrobial susceptibility testing; 11th informational supplement. Approved standard M2-A7 and M7-A5. Natl. Comm. Clin. Lab. Stand., Wayne, PA, USA.
- 23. Oliveira, S.D.; Flores, F.S.; Santos, L.R.; Brandelli, A. (2005). Antimicrobial resistance in Salmonella Enteritidis strains isolated from broiler carcasses, food, human and poultry-related samples. Int. J. Food Microbiol, 97, 297-305.
- 24. Oliveira, S.D.; Rodenbusch, C.R.; Michael, G.; Cardoso, M.I.R.; Canal, C.W.; Brandelli, A. (2003). Detection of virulence genes in Salmonella Enteritidis isolated from different sources. Braz. J. Microbiol, 34, 123-124.
- 25. Peresi, J.T.M.; Almeida, I.A.Z.C.; Lima, S.I.; Marques, D.F.; Rodrigues, E.C.A.; Fernandes, S.A.; Gelli, D.S.; Irino, K. (1998). Surtos de enfermidades transmitidas por alimentos causados por Salmonella Enteritidis. Rev. Saude Publica, 32, 477-483.
- 26. Rademaker, J.L.W.; Bruijn, F.J. (1997). Characterization and classification of microbes by REP-PCR genomic fingerprinting and computer-assisted pattern analysis. In Caetano-Anollés, G, Gresshoff, P. M. (Eds.), DNA markers: protocols, applications, and overviews. J. Wiley Sons, New York, pp.151-171.
- 27. Rasschaert, G.; Houf, K.; Imberechts, H.; Grijspeerdt, K.; De Zutter, L.; Heyndrickx, M. (2005). Comparison of five repetitive-sequence-based PCR typing methods for molecular discrimination of Salmonella enterica isolates. J. Clin. Microbiol., 43, 3615-3623.
- 28. Rodrigue, D.C.; Tauxe, R.V.; Rowe, B. (1990). International increase in Salmonella Enteritidis: a new pandemic? Epidemiol. Infect, 105, 21-27.
- 29. Santos, D.M.S.; Berchieri Junior, A.; Fernandes, S.A.; Tavechio, A.T.; Amaral, L.A. (2000). Salmonella em carcaças de frango congeladas. Pesq. Vet. Bras, 20, 39-42.
- 30. Santos, L.R. (2001). Fagotipagem e análise por RAPD/PCR (DNA Polimórfico Amplificado ao Acaso) de amostras de Salmonella Enteritidis isoladas de materiais de origem avícola e de alimentos e humanos envolvidos em casos de toxinfecções alimentares. M. D. thesis, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil.
- 31. Santos, L.R.; Nascimento, V.P.; Oliveira, S.D.; Rodrigues, D.P.; Reis, E.M.F.; Seki, L.M.; Ribeiro, A.R.; Fernandes, S.A. (2003). Phage types of Salmonella Enteritidis isolated from clinical and food samples, and from broiler carcasses in Southern Brazil. Rev. Inst. Med. Trop. São Paulo, 45, 1-4.
- 32. Tassios, P.T.; Markogiannakis, A.; Vatopoulos, A.C.; Katsanikou, E.; Velonakis, E.N.; Kourea-Kremastinou, J.; Legakis, N.J. (1997). Molecular epidemiology of antibiotic resistance of Salmonella enteritidis during a 7-year period in Greece. J. Clin. Microbiol., 35, 1316-1321.
- 33. Tavechio, A.T.; Fernandes, S.A.; Neves, B.C.; Dias, A.M.G.; Irino, K. (1996). Changing patterns of Salmonella serovars: increase of Salmonella Enteritidis in São Paulo, Brazil. Rev. Inst. Med. Trop. São Paulo, 38, 315-322.
- 34. Tavechio, A.T.; Ghilardi, A.C.; Peresi, J.T.; Fuzihara, T.O.; Yonamine, E.K.; Jakabi, M.; Fernandes, S.A. (2002). Salmonella serotypes isolated from nonhuman sources in São Paulo, Brazil, from 1996 through 2000. J. Food Prot, 65, 1041-1044.
- 35. Thong, K.L.; Ngeow, Y.F.; Altwegg, M.; Navaratnam, P.; Pang, T. (1995). Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J. Clin. Microbiol, 33, 1070-1074.
- 36. Tompkins, L.S.; Troup, N.; Labaigne-Roussel, A.; Cohen, M. (1986). Cloned random chromosomal sequences as probes to identify Salmonella species. J. Infect. Dis., 154, 156-162.
- 37. Van Lith, L.A.J.T.; Aarts, H.J.M. (1994). Polymerase chain reaction identification of Salmonella serotypes. Lett. Appl. Microbiol, 19, 273-276.
- 38. Versalovic, J.; Koeuth, T.; Lupski, J.R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res, 19, 6823-6831.
- 39. Versalovic, J.; Schneider, M.; Bruijn, F.J.; Lupski, J.R. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol, 5, 25-40.
- 40. Weigel, R.M.; Qiao, B.; Barber, D.A.; Teferedegne, N.B.; Kocherginskaya, S.; White, B.A.; Isaacson, R.E. (2001). Identification of patterns of transmission of Salmonella within swine production systems using pulsed field gel electrophoresis (PFGE) and repetitive sequence polymerase chain reaction (REP-PCR): a quantitative analysis. Berl. Munch. Tierarztl. Wochenschr, 114, 397-400.
Publication Dates
-
Publication in this collection
28 Jan 2008 -
Date of issue
Dec 2007
History
-
Accepted
20 Sept 2007 -
Reviewed
19 May 2007 -
Received
28 Feb 2007