Abstracts
The main objective of this work was to develop a PCR protocol for the identification of Fusarium graminearum, based on a pair of primers targeted to a segment of the 3' coding region of the gaoA gene that codes for the enzyme galactose oxidase (GO). This region has low homology with the same region of GO genes from other fungi. Genomic DNA from 17 strains of Fusarium spp. isolated from diseased cereals, from several other Fusarium species, and from other fungi genera was analyzed in a PCR assay using this primer set. The 17 strains of Fusarium spp. were also analyzed for the GO enzyme production in submerse fermentation in a new formulated liquid medium. All strains that were morphologically and molecularly identified as F. graminearum were able to secrete the enzyme and had a positive result in the used PCR protocol. No DNA fragment was amplified using genomic DNA from other Fusarium species and species of other fungi genera. The results suggest that the proposed PCR protocol is specific and can be considered as a new molecular tool for the identification of F. graminearum. In addition, the new formulated medium is a cheap alternative for screening for GO screening production by F. graminearum.
Fusarium graminearum; galactose oxidase; gaoA gene; molecular identification
O principal objetivo deste trabalho foi desenvolver um novo protocolo de PCR para identificação de isolados de Fusarium graminearum, baseado no uso de um par de iniciadores direcionado para um segmento da região 3' codificadora do gene gaoA que codifica a enzima galactose oxidase (GO). Esta região possui baixa homologia com a mesma região de genes da GO de outros fungos. O DNA genômico de 17 cepas de Fusarium spp. isoladas de cereais infectados com sintomas, de vários outras espécies de Fusarium e de outros gêneros de fungos foi analisado em um protocolo de PCR utilizando os iniciadores desenhados. Os 17 isolados de Fusarium spp. também foram analisados para a produção da enzima GO em fermentação submersa em um novo meio líquido. Todas as cepas que foram morfologicamente e molecularmente identificadas como F. graminearum foram capazes de secretar a enzima e tiveram um resultado positivo no protocolo de PCR, utilizando os iniciadores direcionados para o gene gaoA. Nenhum fragmento de DNA foi amplificado quando foi utilizado o DNA genômico de várias outras espécies de Fusarium e de espécies de outros gêneros de fungos. Os resultados sugerem que o protocolo de PCR gerado é específico e pode ser considerado como uma nova ferramenta molecular para a identificação de cepas de F. graminearum. Além disso, o meio líquido formulado é uma alternativa barata para a avaliação da produção de GO por F. graminearum.
Fusarium graminearum; galactose oxidase; gene gaoA; identificação molecular
INDUSTRIAL MICROBIOLOGY
A new PCR approach for the identification of Fusarium graminearum
Um novo protocolo de PCR para a identificação de Fusarium graminearum
Gleison Ricardo de BiazioI; Gabriela Guimarães Sousa LeiteI; Dauri José TessmannII; Ione Parra Barbosa-TessmannI,* * Corresponding Author. Mailing address: Universidade Estadual de Maringá - Departamento de Bioquímica, Av. Colombo, 5790. Bloco I89 Sala 11A 87020-900 Maringá PR. Tel.: 44 3261 4808 Fax 44 3261 4714. E-mail: ipbtessmann@uem.br
IDepartamento de Bioquímica, Universidade Estadual de Maringá, Maringá, PR, Brasil
IIDepartamento de Agronomia, Universidade Estadual de Maringá, Maringá, PR, Brasil
ABSTRACT
The main objective of this work was to develop a PCR protocol for the identification of Fusarium graminearum, based on a pair of primers targeted to a segment of the 3' coding region of the gaoA gene that codes for the enzyme galactose oxidase (GO). This region has low homology with the same region of GO genes from other fungi. Genomic DNA from 17 strains of Fusarium spp. isolated from diseased cereals, from several other Fusarium species, and from other fungi genera was analyzed in a PCR assay using this primer set. The 17 strains of Fusarium spp. were also analyzed for the GO enzyme production in submerse fermentation in a new formulated liquid medium. All strains that were morphologically and molecularly identified as F. graminearum were able to secrete the enzyme and had a positive result in the used PCR protocol. No DNA fragment was amplified using genomic DNA from other Fusarium species and species of other fungi genera. The results suggest that the proposed PCR protocol is specific and can be considered as a new molecular tool for the identification of F. graminearum. In addition, the new formulated medium is a cheap alternative for screening for GO screening production by F. graminearum.
Key-words: Fusarium graminearum, galactose oxidase, gaoA gene, molecular identification.
RESUMO
O principal objetivo deste trabalho foi desenvolver um novo protocolo de PCR para identificação de isolados de Fusarium graminearum, baseado no uso de um par de iniciadores direcionado para um segmento da região 3' codificadora do gene gaoA que codifica a enzima galactose oxidase (GO). Esta região possui baixa homologia com a mesma região de genes da GO de outros fungos. O DNA genômico de 17 cepas de Fusarium spp. isoladas de cereais infectados com sintomas, de vários outras espécies de Fusarium e de outros gêneros de fungos foi analisado em um protocolo de PCR utilizando os iniciadores desenhados. Os 17 isolados de Fusarium spp. também foram analisados para a produção da enzima GO em fermentação submersa em um novo meio líquido. Todas as cepas que foram morfologicamente e molecularmente identificadas como F. graminearum foram capazes de secretar a enzima e tiveram um resultado positivo no protocolo de PCR, utilizando os iniciadores direcionados para o gene gaoA. Nenhum fragmento de DNA foi amplificado quando foi utilizado o DNA genômico de várias outras espécies de Fusarium e de espécies de outros gêneros de fungos. Os resultados sugerem que o protocolo de PCR gerado é específico e pode ser considerado como uma nova ferramenta molecular para a identificação de cepas de F. graminearum. Além disso, o meio líquido formulado é uma alternativa barata para a avaliação da produção de GO por F. graminearum.
Palavras-chave: Fusarium graminearum, galactose oxidase, gene gaoA, identificação molecular.
INTRODUCTION
F. graminearum has been described as the major causal agent of the scab or fusarium head blight on wheat and barley in several regions of the world, including Brazil (9,17,22). This disease is recognized as one of the most destructive pathology of small cereal crops, reducing grain yield and quality. F. graminearum is also a known producer of mycotoxins, mainly of deoxynivalenol and zearalenone, what poses threats to human and animal health and food safety (11,22).
Traditional diagnostic methods for detection and identification of F. graminearum in culture or in infected grains are based on micro and macro morphological features. This process is time consuming, requires training, and it can often be difficult to distinguish between similar species. Molecular methods, more sensitive and faster are also employed to the specific identification of Fusarium species. There are reports of the use of PCR with primers targeted to the internal transcribed sequence (ITS) of the ribosomal DNA (24) for the detection and identification of F. graminearum. However, sequences in the ITS regions have shown to be highly variable in fusaria (20). There also is a report of PCR primers targeted to the junction promoter/5'-coding region of the gaoA gene, which codes for the enzyme galactose oxidase (GO), for the identification of F. graminearum (19). But, the authors of that primer set have reported that it failed to identify one F. graminearum strain that was a GO producer (19).
The monomeric enzyme galactose oxidase (D-galactose: O2 oxidorredutase, EC. 1.1.3.9) is a copper enzyme that catalyzes the oxidation of primary alcohols to its respective aldehydes, with concomitant reduction of O2 to H2O2 (23,30). This enzyme has several biotechnological applications including: lactose and galactose assays in dairy industry and in clinical laboratory (1,13), carbohydrate synthesis (16), biotransformation of glycobiopolymers (6), histochemistry studies (25), and colon cancer diagnosis (26).
Extra cellular production of GO has been shown for F. graminearum (3,5,7), Gibberella fujikuroi (2), Fusarium moniliforme f. sp. subglutinans (5), and Fusarium acuminatum (5). However, most of the studies and the industrial production of GO use the isolate F. graminearum NRRL 2903, that was initially classified as Dactylium dendroides (3,19,21). The gaoA gene has no intron and has been cloned for this strain (18).
Problems associated with the use of GO regards the availability of sufficiently pure enzyme. The purification procedures are laborious and the commercial preparations are very expensive (16). Because of these reasons, the applications of GO would be beneficial by the finding of new producer isolates that could be able to produce high levels of the enzyme. Traditional methods for screening GO production are based on submerse fermentation and search for enzyme activity in the liquid medium (5,7).
Because it would be interesting the development of a new molecular assay for the identification of F. graminearum, the main objective of this study was to develop a new PCR protocol using a primer set targeted to the internal 3'-coding region of the F. graminearum gaoA gene. If proven specific, this method could be used for F. graminearum identification for fusarium head blight diagnosis and food quality assessment. Another objective of this work was to develop a new medium for GO screening production.
MATERIAL AND METHODS
Microorganisms and maintenance
Seventeen strains of Fusarium spp. (Table 1) were isolated from diseased wheat and triticale from Southern Brazil. The method for isolation, the geographic origin, the host, and the plant organ where they were isolated were described elsewhere (4,10,11). The isolates UEM 67 and UEM 68 were isolated in the city of Campo Mourão, Paraná state, from wheat seed. The F. graminearum original GO producer strain (GOPS) (3) and two other GO Fusarium spp. producers, F. acuminatum UnB 356 (5) and F. moniliforme f. sp. subglutinans UnB 379 (5), were used as references. All isolates are being maintained in potato dextrose agar (PDA) slants with trimestral transfer and in stocks under mineral oil.
Culture conditions for enzyme production
The liquid medium used for submerse fermentation was developed based on other media described in the literature (15,27,28). This medium was prepared in three separate solutions: solution A: 62 mM Na2HPO4, 62 mM KH2PO4, 13 mM (NH4)NO3, 15 mM (NH4)2SO4, 15 mM NaOH, 14 mM KOH, 0.1% yeast extract, pH 7.0; solution B: 1.6 mM MgSO4·7H2O, 11.8 µM MnSO4·H2O, 10 µM CuSO4·5H2O; and solution C: 1% glucose. The indicated concentrations are the ones in the final medium. Solutions A and B were sterilized by autoclaving, for 20 minutes. Solution C was sterilized in fluent vapor, for 20 minutes. Before use, the three solutions were aseptically combined: 22.5 ml of solution A, 0.5 ml of solution B, and 2.0 ml of solution C. A fragment of a fresh PDA culture (1 cm3) was used as inoculum for 125 ml Erlenmeyer culture flasks containing 25 ml of the liquid medium. Culture involved an initial 4 day inoculum growth on a rotary shaker at 100 r.p.m., in dark, at 25ºC, followed by homogenization using passage first through a sterile sieve and then through a sterile 18-gauge needle attached to a sterile syringe. The homogenized was used as inoculum (2% v/v) to new 125 ml culture flasks, containing 25 ml of the same liquid medium, which were grown in the same conditions, for 72 hours, and then filtered through filter paper. The culture filtrates were used for the enzymatic analysis. The mycelia biomass was determined after drying the mycelia at 50ºC for at least 24 hours.
Enzyme assay
GO activity was assayed by the peroxidase/o-dianisidine colorimetric method (3,28), based on the estimation of the colored oxidized o-dianisidine. Filtrate (0.5 ml pure or diluted); 1.4 ml of the reactive mixture [50 mM phosphate buffer, pH 7.0; 0.2 mg/ml o-dianisidina (Sigma D-3252; previously dissolved in methanol 2 mg/ml); 0.04 mg/ml (6.0 U/ml) peroxidase (Sigma P-8125)]; and 0.1 ml of 0.5 M D-(+)-galactose were mixed and incubated for 10 minutes at 30ºC. The brown colored product was read at 460 nm. For these conditions, one enzymatic unit corresponded to an absorbance of 1.0. Culture medium filtrates that resulted in no activity at the 10 minutes reaction in the enzyme assay were incubated for extra 20 minutes and the absorbance was read again.
DNA Extraction
Mycelia of the 17 isolates of Fusarium spp. and of the reference isolates were obtained in liquid culture, as described above. Mycelia of the other Fusarium species isolates and of the other fungi genera were obtained by transferring a fragment of a fresh PDA slant culture to 25 ml of the liquid medium described above or to 25 ml of liquid potato dextrose medium, respectively, in 125 ml Erlenmeyer flasks and culturing for three 3 days, in the conditions described above. Genomic DNA was extracted using the protocol described by Koenig et al. (14) and modified as follows. The mycelia mass obtained in liquid culture was collected by filtration in sterilized gauze, macerated in liquid nitrogen, and transferred to microcentrifuge tubes. The extraction buffer was added to the macerated mycelia in the proportion of 700 µl to each 300 µl of macerated mycelia. The extraction buffer contained, in the proportion of 1.0/1.0/0.4, nuclear lyses buffer (0.2 M Tris, pH 7.5; 50 mM EDTA; 2% (w/v) cetyltrimethylammonium bromide; pH 7.5), DNA isolation buffer (0.35 M sorbitol; 0.1 M Tris, pH 7.5; 5 mM EDTA; pH 7.5), and 5% Sarkosyl. The extraction buffer was combined right before use and was then added of 3.8 mg/ml of sodium bisulfite. The tubes were incubated in a dry bath at 65ºC for 60 minutes. After that, 500 µl of a mixture of chloroform:isoamylic alcohol (24:1) were added. The tubes were centrifuged at room temperature (12.000g, 10 minutes) and the supernatant fraction was transferred to clean tubes. Samples were treated with five µl of RNAse A (20 mg/ml), for 30 minutes, at 37ºC, and after with 5 µl of proteinase K (20 mg/ml), for 30 minutes, at 56ºC. The DNA was then precipitated with equal volume of isopropanol and overnight incubation at -20ºC. The precipitated DNA was collected by centrifugation at room temperature (12.000g, 10 minutes) and the DNA pellet was washed three times with cold 70% ethanol. The final DNA pellet was dried at room temperature and ressuspended with 50 µl of TE buffer (10 mM Tris, pH 8.0; 1 mM EDTA). The whole process was carried out under sterile conditions. The DNA was quantified in a spectrophotometer at 260 nm. The DNA final concentration was adjusted to 100 ng/µl in TE buffer.
PCR Analysis
For primer designing, GO coding genes from F. graminearum (gaoA gene) (8,18) (GenBank AN M86819), from Fusarium venenatum (12), from Aspergillus oryzae (GenBank AN BD160924), and from Fusarium sporotrichioides (GenBank AN BI201662 and GenBank AN BI190798) were aligned using the ClustalW program (Fig. 1a). A region of low homology was chosen for primer designing. The primer set (GOFW 5'-ACCTCTGTTGTTCTTCCAGACGG-3'and GORV 5'-CTGGTCAGTATTAACCGTGTGTG-3') were designed to amplify a 435 pb DNA fragment of a internal 3'-coding region of the gene (Fig. 1b), based on the sequence published by McPherson et al. (18) and Firbank et al. (8). Primers used for F. graminearum specific identification (UBC85F410-UBC85R410) were the ones described by Schiling et al. (24). All genomic DNAs used in this work were tested for PCR amplification suitability using the primers ITS4 and ITS5 targeted to the ITS-5.8 S rRNA region as described by White et al. (29). A DNA fragment of approximately 550 bp was amplified from all genomic DNAs using the ITS4 and ITS5 primers (not shown). The amplification reactions were performed in a termocycler Techne TC-312 (England) in PCR tubes containing 25 µL of the following reaction mixture: 50 mM KCl; 10 mM Tris, pH 7.5; 1.5 mM MgCl2, 1.5 U of Platinum Taq DNA polymerase (Invitrogen); 0.2 mM of each dNTP (Invitrogen); 25 pmol of each primer (forward and reverse), and 20-400 ng of the DNA sample. The PCR reaction consisted of 25 cycles of 1 minute and 30 seconds at 94ºC, 1 minute and 30 seconds at 55ºC for the GO and UBC85 primers and at 50oC for the ITS primers, and 2 minutes at 72ºC. Previously to the cycles, the samples were heated for 5 minute at 94ºC, and after the cycles the samples were incubated for 10 minutes at 72ºC and frozen at -20ºC until use. Positive control was the DNA from the isolate F. graminearum GOPS. Negative controls (no DNA template) were used in each experiment to test for the presence of DNA contamination of reagents and reaction mixtures. Ten microliters of the PCR reaction were analyzed in an agarose gel containing ethidium bromide (0.25 µg/ml). Molecular weight markers were the 100 bp markers from Promega or Invitrogen, USA. The PCR products were visualized and photographed under UV light.
RESULTS
The results of the enzyme production by the isolates are summarized in Table 1. No enzyme activity was detected in the filtrates of the isolates UEM 32, UEM 68, and UEM 69, even in the 30 min enzyme assay. Some isolates produced a good level of the enzyme, but no isolate could produce levels of GO higher than the ones produced by the reference isolate (GOPS).
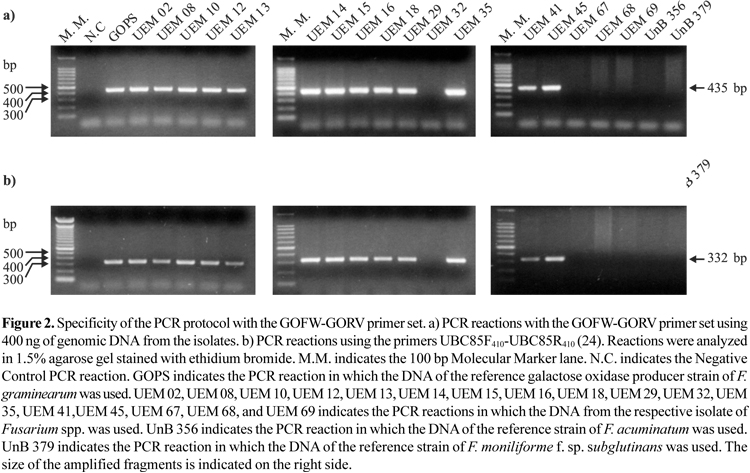
PCR analysis with the GOFW and GORV primers revealed amplification of a single DNA fragment from purified genomic DNA of all F. graminearum strains (Fig. 2a, Table 1). The obtained fragment size was in good agreement with the theoretical size of the fragment (435 bp) expected from the gaoA gene sequence data (18). The GO primers had the same specificity as the F. graminearum specific primers UBC85 (Fig. 2b, Table 1), which could amplify a 332 bp fragment from the same isolates. The used PCR protocol and primers could not amplify any fragment, specific and non specific, using DNA template from other species of Fusarium and from species of other fungi genera (Table 1).
To test the method sensitivity, the DNA of the GOPS isolate was ten-fold serially diluted and tested in the PCR protocol (Fig. 3). The PCR method could amplify a DNA fragment with concentrations of DNA as low as 4 ng.
DISCUSSION
Markus et al. (15) optimized the production of galactose oxidase by F. graminearum in shake culture flasks. They described a liquid culture medium composed by nitrate and ammonia as the nitrogen source, yeast extract, glucose as the carbon source, and mineral ions. They also reported that thiamine could substitute the yeast extract with no loss in enzyme production, and that sorbose could replace glucose with an increment in enzyme production. Subsequently, Shatzman and Kosman (27) reported the influence of medium pH on enzyme production. More recently, Tressel and Kosman (28) described a medium containing nitrate and ammonia as the nitrogen source, thiamine as the only vitamin, sorbose as the carbon source, mineral ions, and having a better buffering system for pH 7.0. Sorbose is an expensive carbohydrate and some of the strains used in this work could not grow well in the medium reported by Tressel and Kosman (28). Because of this, a combined and new medium was formulated. This new medium has the buffering system of the medium developed by Tressel and Kosman (28) and the other components used in the medium described by Markus et al. (15). All used microorganisms could grow well in this new medium and it represents a cheap alternative for F. graminearum GO screening production in submerse fermentation.
All isolates that could pigment the PDA medium with a red pigment and that did not produce spores in liquid culture, similar to the reference F. graminearum strain GOPS (Table 1), could produce the enzyme and had a DNA fragment of the gaoA gene amplified in the PCR reaction. In addition, all of these isolates were molecularly identified as F. graminearum with the UBC85 primers (Table 1, Fig. 2b). These results are in agreement with previous screening studies, which evidenced that production of galactose oxidase is much more abundant among F. graminearum strains (5,7).
The primers targeted to the gaoA gene used in this study appears to be specific for F. graminearum, since they did not amplify any DNA fragment, specific or non specific, when the genomic DNA of other GO secreting Fusarium species was used in the PCR protocol, including the genomic DNA of the GO producing reference isolates F. acuminatum and F. moniliforme f. sp. subglutinans (Fig. 2a). This may indicate differences among the GO gene of those Fusarium species with the gaoA gene from F. graminearum, within the primers localization. Considering this, the isolate UEM 67 that was not morphologically nor molecularly identified as F. graminearum, with the UBC85 primers and with the GO primers (Table 1; Fig. 2a), but produced the enzyme in a very low level could be one of these reference species, probably F. moniliforme f. sp. subglutinans, because of the spore production in the liquid culture medium.
The designed pair of primers targeted to the GO gene could not amplify any fragment of DNA, specific or non specific, when genomic DNA from other fungi genera was used in the established PCR protocol (Table 1). This certifies the specificity of the generated method.
Taking into consideration the specificity of the GO primers PCR protocol, the generated methodology could be considered as a new molecular tool for F. graminearum identification. Its use for the molecular identification of F. graminearum represents an advance for head blight diagnosis and food safety assessment.
ACKNOWLEDGEMENTS
This research was supported by: the International Foundation for Science, Stockholm, Sweden, and The Organization for the Prohibition of Chemical Weapons (OPCW), THE HAGUE, Netherlands, through a grant to Dr Ione Parra Barbosa Tessmann. This research was also supported by the Fundação Araucária (Secretaria da Ciência e Tecnologia do Estado do Paraná, Brazil). The authors wish to thank Dr. Carlos Kemmelemeir and Dr. José Carmine Dianese for the kind donation of some of the isolates.
Submitted: October 17, 2007; Returned to authors for corrections: December 24, 2007; Approved: July 13, 2008
- 1. Adányi, N.; Szabó, E.E.; Váradi, M. (1999). Multi-enzyme biosensors with amperometric detection for determination of lactose in milk and dairy products. Eur. Food Res. Technol., 209, 220-226.
- 2. Aisaka, K.; Terada, O. (1981). Production of galactose oxidase by Gibberella fujikuroi Agric. Biol. Chem., 45, 2311-2316.
- 3. Amaral, D.; Kelly-Falcoz, F.; Horecker, B.L. (1966). Galactose oxidase of Polyporus circinatus Meth. Enzymol., 9, 87-92.
- 4. Angelotti, F.; Tessmann, D.J.; Alves, T.C.A.; Vida, J.B.; Jaccoud Filho, D.S.; Harakava, R. (2006). Caracterização morfológica e identificação molecular de isolados de Fusarium graminearum associados à giberela do trigo e triticale no sul do Brasil. Summa Phytopathologica, 32, 177-179.
- 5. Barbosa-Tessmann, I.P.; Da Silva, D.A.; Peralta, R.M.; Kemmelmeier, C. (2001). A new species of Fusarium producer of galactose oxidase. J. Basic Microbiol., 41, 143-148.
- 6. Chiu, C.W.; Jeffcoat, R.; Henley, M.; Peek, L. (1996). Aldehyde cationic derivatives of galactose containing polysaccharides used as paper strength additives. US Pat, 5,554,745.
- 7. Dias, D.; Kemmelmeier, C. (1987). Ocorrência da galactose oxidase em Fusarium graminearum Rev. Microbiol, 18, 276-278.
- 8. Firbank, S.J.; Rogers, M.S.; Wilmot, C.M.; Dooley, D.M.; Halcrow, M.A.; Knowles, P.F.; McPherson, M.J.; Phillips, S.E.V. (2001). Crystal structure of galactose oxidase: an unusual self-processing enzyme. Proc. Natl. Acad. Sci. USA, 98, 12932-12937.
- 9. Gale, L.R. (2003). Population biology of Fusarium species causing head blight of grain crops. In: Leonard, K.J., Bushnell, W.R. (eds). Fusarium Head Blight of Wheat and Barley APS Press, St. Paul, USA, pp. 120-143.
- 10. Gasparotto, E.P.L.; Abrão, S.C.C.; Inagaki, S.Y.; Tessmann, D.J.; Kemmelemeir, C.; Barbosa-Tessmann, I.P. (2006). Production and characterization of galactose oxidase produced by four isolates of Fusarium graminearum Braz. Arch. Biol. Technol., 49, 557-564.
- 11. Geraldo, M.R.F.; Tessmann, D.J.; Kemmelmeier, C. (2006). Production of mycotoxins by Fusarium graminearum isolated from small cereals (wheat, triticale and barley) affected with scab disease in southern Brazil. Braz. J. Microbiol., 37, 58-63.
- 12. Golightly, E.; Berka, R.M.; Rey, M.W. (2001). Polypeptides having galactose oxidase activity and nucleic acids encoding same. US Pat., 6,277,612.
- 13. Karube, I.; Kimura, J.; Yokoyama, K.; Tamiya, E. (1990). Integrated microbiosensors for clinical diagnosis. Ann. N. Y. Acad. Sci., 613, 385-389.
- 14. Koenig, R.L.; Ploetz, R.C.; Kistler, H.C. (1997). Fusarium oxysporum f.sp. cubense consists of a small number of divergent and globally distributed clonal lineages. Phytopathol., 87, 915-923.
- 15. Markus, Z.; Miller, G.; Avigad, G. (1965). Effect of culture conditions on the production of D-galactose oxidase by Dactylium dendroides Appl. Microbiol., 13, 686-693.
- 16. Mazur, A.W. (1991). Galactose oxidase. Selected properties and synthetic applications. ACS Symposium Series, 466, 99-110.
- 17. McMullen, M.; Jones, R.; Gallenberg, D. (1997). Scab of wheat and barley: A re-emerging disease of devastating impact. Plant Dis., 81, 1340-1348.
- 18. McPherson, M.J.; Ögel, Z.B.; Stevens, C.; Yadav, K.D.S.; Keen, J.N.; Knowles, P.F. (1992). Galactose oxidase of Dactylium dendroides Gene cloning and sequence analysis. J. Biol. Chem., 267, 8146-8152.
- 19. Niessen, M.L.; Vogel, R.F. (1997). Specific identification of Fusarium graminearum by PCR with gaoA targeted primers. System. Appl. Microbiol., 20, 111-113.
- 20. O'Donnell, K. (1992). Ribossomal DNA internal transcribed spacers are highly diverged in the phytopathogenic ascomycete Fusarium sambucinum (Gibberella pulicaris). Curr. Genet., 22, 213-220.
- 21. Ögel, Z.B.; Brayford, D.; McPherson, M.J. (1994). Cellulose-triggered sporulation in the galactose oxidase-producing fungus Cladobotryum (Dactylium) dendroides NRRL 2903 and its re-identification as a species of Fusarium Mycol. Res., 98, 474-480.
- 22. Parry, D.W.; Jenkinson, P.; McLeod, L. (1995). Fusarium ear blight (scab) in small-grain cereals-A review. Plant Pathol, 44, 207-238.
- 23. Rogers, M.S.; Dooley, D.M. (2003). Copper-tyrosyl radical enzymes. Curr. Op. Chem. Biol., 7, 189-196.
- 24. Schiling, A.G.; Möller, E.M.; Geiger, H.H. (1996). Polymerase Chain Reaction-Based Assays for Species-Specific Detection of Fusarium culmorum, F. graminearum, and F. avenaceum Phytopathol., 86, 515-522.
- 25. Schulte, B.A.; Spicer, S.S. (1983). Light microscopic histochemical detection of sugar residues in secretory glycoproteins of rodent and human tracheal glands with lectin-horseradish peroxidase conjugates and the galactose oxidase-Schif sequence. J. Histochem. Cytochem., 31, 391-403.
- 26. Shamsuddin, A.M. (1996). A simple test for cancer screening. Anticancer Res., 16, 2193-2200.
- 27. Shatzman, A.R.; Kosman, D.J. (1977). Regulation of galactose oxidase synthesis and secretion in Dactylium dendroides: effects of pH and culture density. J. Bacteriol., 130: 455-463.
- 28. Tressel, P.S.; Kosman, D.J. (1982). Galactose oxidase from Dactylium dendroides Meth. Enzymol., 89, 163-171.
- 29. White, T.J.; Bruns, T.; Lee, S.; Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, H., Sninsky, J.J., White, T.J. (eds). PCR Protocols, a Guide to Methods and Applications Academic Press Inc, San Diego, USA, pp. 315-322.
- 30. Whittaker, J.W. (2002). Galactose oxidase. Adv. Prot. Chem., 60, 1-49.
Publication Dates
-
Publication in this collection
02 Oct 2008 -
Date of issue
Sept 2008
History
-
Accepted
13 July 2008 -
Received
17 Oct 2007