Abstracts
In the last decades, coagulase-negative staphylococci (CoNS), especially Staphylococcus epidermidis have become an important cause of bloodstream infections. In addition, rates of methicillin-resistance among CoNS have increased substantially, leading to the use of glicopeptides for therapy. The objective of this study was to evaluate eleven consecutives clinically relevant cases of oxacillin-resistant CoNS bacteremia in a general hospital localized in São Paulo city, Brazil. Five different species were identified by different phenotypic methods, including S. epidermidis (5), S. haemolyticus (3), S. hominis (1), S. warneri (1) and S. cohnii subsp urealyticus (1). A variety of Pulsed Field Gel Electrophoresis profiles was observed by macrorestriction DNA analysis in S. epidermidis isolates, but two of three S. haemolyticus isolates presented the same profile. These data indicated the heterogeneity of the CoNS isolates, suggesting that horizontal dissemination of these microorganisms in the investigated hospital was not frequent. One S. epidermidis and one S. haemolyticus isolates were resistant to teicoplanin and susceptible to vancomycin. The selective pressure due to the use of teicoplanin in this hospital is relevant.
Coagulase-negative staphylococci; bacteremia; oxacillin; PFGE
Staphylococcus coagulase negativos (SCoN), especialmente Staphylococcus epidermidis tem se tornado causa importante de infecções da corrente circulatória nas últimas décadas. Além disso, percentuais de resistência a meticilina entre os SCoN têm aumentado significativamente, levando ao uso de glicopeptídeos nestes pacientes. O objetivo deste estudo foi avaliar onze casos consecutivos de bacteremia clinicamente relevantes por SCoN oxacilina resistentes em um hospital localizado na cidade de São Paulo, Brasil. Cinco diferentes espécies foram identificadas por diferentes métodos fenotípicos, incluindo S. epidermidis (5), S. haemolyticus (3), S. hominis (1), S. warneri (1) e S. cohnii subsp urealyticus (1). Diferentes perfis eletroforéticos obtidos pela técnica de "Pulsed Field Gel Electrophoresis" foram observados na análise da macrorestrição do DNA nos isolados de S. epidermidis, mas dois dos três isolados de S. haemolyticus apresentaram o mesmo perfil. Esses dados indicam uma heterogeneidade nos isolados SCoN, sugerindo que a disseminação horizontal no hospital investigado não é freqüente. Um isolado de S. epidermidis e um de S. haemolyticus foram resistentes à teicoplanina e sensíveis à vancomicina. Observa-se a relevância da pressão seletiva pelo uso de teicoplanina nos pacientes deste hospital.
Staphylococcus spp. coagulase negativo; bacteremia; oxacilina; PFGE
MEDICAL MICROBIOLOGY
Oxacilin-resistant Coagulase-negative staphylococci (CoNS) bacteremia in a general hospital at São Paulo city, Brasil
Bacteremias por Staphylococcus coagulase negativos oxacilina resistentes em um hospital na cidade de São Paulo, Brasil
P.A. d'AzevedoI,II; C. SecchiII; A.L.S.AntunesII; T.SalesIII; F.M.SilvaI; R.TranchesiIII; A.C.C.PignatariI,III
ILaboratório Especial de Microbiologia Clínica, Universidade Federal de São Paulo, São Paulo, SP, Brasil
IILaboratório de Cocos Gram Positivos, Universidade Federal de Ciências da Saúde de Porto Alegre, Porto Alegre, RS, Brasil
IIIHospital 9 de Julho, São Paulo, SP, Brasil
ABSTRACT
In the last decades, coagulase-negative staphylococci (CoNS), especially Staphylococcus epidermidis have become an important cause of bloodstream infections. In addition, rates of methicillin-resistance among CoNS have increased substantially, leading to the use of glicopeptides for therapy. The objective of this study was to evaluate eleven consecutives clinically relevant cases of oxacillin-resistant CoNS bacteremia in a general hospital localized in São Paulo city, Brazil. Five different species were identified by different phenotypic methods, including S. epidermidis (5), S. haemolyticus (3), S. hominis (1), S. warneri (1) and S. cohnii subsp urealyticus (1). A variety of Pulsed Field Gel Electrophoresis profiles was observed by macrorestriction DNA analysis in S. epidermidis isolates, but two of three S. haemolyticus isolates presented the same profile. These data indicated the heterogeneity of the CoNS isolates, suggesting that horizontal dissemination of these microorganisms in the investigated hospital was not frequent. One S. epidermidis and one S. haemolyticus isolates were resistant to teicoplanin and susceptible to vancomycin. The selective pressure due to the use of teicoplanin in this hospital is relevant.
Key-words: Coagulase-negative staphylococci; bacteremia; oxacillin; PFGE.
RESUMO
Staphylococcus coagulase negativos (SCoN), especialmente Staphylococcus epidermidis tem se tornado causa importante de infecções da corrente circulatória nas últimas décadas. Além disso, percentuais de resistência a meticilina entre os SCoN têm aumentado significativamente, levando ao uso de glicopeptídeos nestes pacientes. O objetivo deste estudo foi avaliar onze casos consecutivos de bacteremia clinicamente relevantes por SCoN oxacilina resistentes em um hospital localizado na cidade de São Paulo, Brasil. Cinco diferentes espécies foram identificadas por diferentes métodos fenotípicos, incluindo S. epidermidis (5), S. haemolyticus (3), S. hominis (1), S. warneri (1) e S. cohnii subsp urealyticus (1). Diferentes perfis eletroforéticos obtidos pela técnica de "Pulsed Field Gel Electrophoresis" foram observados na análise da macrorestrição do DNA nos isolados de S. epidermidis, mas dois dos três isolados de S. haemolyticus apresentaram o mesmo perfil. Esses dados indicam uma heterogeneidade nos isolados SCoN, sugerindo que a disseminação horizontal no hospital investigado não é freqüente. Um isolado de S. epidermidis e um de S. haemolyticus foram resistentes à teicoplanina e sensíveis à vancomicina. Observa-se a relevância da pressão seletiva pelo uso de teicoplanina nos pacientes deste hospital.
Palavras-chave:Staphylococcus spp. coagulase negativo; bacteremia; oxacilina; PFGE.
INTRODUCTION
Coagulase-negative staphylococci (CoNS) are major causes of nosocomial bloodstream infection and responsible for high morbidity and mortality rates, mainly in hospitalized patients (9). Members of the genera Staphylococcus are catalase-positive, gram-positive cocci, coagulase-negative, aerobes and, when present in human infections, can present multiresistant profiles (8,20). These strains may constitute a dangerous reservoir of resistance genes in a hospital (20).
Staphylococci generally present a benign or symbiotic relationship with their host. However, they may become pathogens when entering the host tissue through break of the cutaneous barrier, inoculation by needles or implantation of medical devices (5). It is increasingly important to accurately identify CoNS isolates to the species level in order to determine the clinical significance of these bacteria, the proper epidemiological surveillance, and the management of patients infected with CoNS in case of relapse (15).
A substantial increase in the frequency of oxacillin-resistance (methicillin-resistant) in CoNS isolates has occurred over the last decades (4). Between 50% and 80%, depending on the species, are mec A positive or oxacillin resistant (1,6).
According to the results of the SENTRY antimicrobial surveillance program, carried out with Brazilian bloodstream isolates over a five-year period from 1997 to 2001, the oxacillin susceptibility in Staphylococcus aureus was 68.2% and 19.2% in CoNS (17).
Staphylococcus epidermidis and Staphylococcus haemolyticus are the most frequent species in nosocomial infections, and the frequency of oxacillin resistance is higher in CoNS clinical isolates (3). S. haemolyticus have been reported to show multiple resistance to antimicrobials and quite frequently clinical isolates present with reduced susceptibility or are resistant to teicoplanin (16).
Vancomycin is usually considered the treatment of choice for infections caused by these microorganisms. However, due to the emergence of vancomycin-resistant enterococci (9) and vancomycin-resistant staphylococci (19), reduction in the use of this drug has been recommended (20). A few reports have shown that the mechanism of glycopeptide resistance in S. epidermidis, S. haemolyticus and S. hominis is similar to that described in VISA and hetero-VISA strains (13). The objective of this study was to evaluate eleven consecutive clinically relevant cases of oxacillin-resistant CoNS bacteremia in a general hospital where therapy with the glycopeptide teicoplanin is broadly utilized.
MATERIAL AND METHODS
Bacterial isolates
The study was carried out with eleven consecutive bloodstream CoNS isolates, obtained between June and July 2005 from patients at 9 de Julho Hospital, a 250 beds general hospital localized in the city of São Paulo, Brazil. According to the CDC criteria, these isolates were considered clinically relevant by the National Nosocomial Infections Surveillance Committee of the hospital (12).
Identification
Staphylococci identification was carried out by test for oxidation-fermentation, coagulase (Laborclin, Brazil), catalase, alkaline phosphatase (Sigma-Aldrich, Germany), ornithine (Merck, Germany), urease (Oxoid, UK), PYR (pyrrolidinyl-β-naphthylamide hydrolysis, Probac do Brasil, Brazil), hemolysis in sheep blood agar, acid production from trehalose (Sigma-Aldrich, Germany), mannitol (Nuclear, Brazil), mannose (Vetec, Brazil), sucrose (Reagen, Brazil), maltose (Sigma-Aldrich, Germany), lactose (Difco , USA), cellobiose (Sigma-Aldrich, Germany) and anaerobic growth in thioglicolate (Merck, Germany). Susceptibility to novobiocin (Oxoid, UK), polymyxin B (Oxoid, UK), bacitracin (CECON, Brazil), desferrioxamine (Ciba Geigy, Switzerland) and fosfomycin (Oxoid,UK) was also determined. Isolates were kept frozen at -20ºC in Skim Milk (Difco, USA). Bacteria to be tested were suspended in 0,5 ml of saline to a McFarland standard and 50 ml were added to each sugar carbohydrate tube. The acid production from carbohydrates was evaluated after 24, 48 and 72hs of incubation at 35-37ºC. The final evaluation was at the 7th day. The phenotypic tests were accomplished in parallel with a positive control (S. epidermidis ATCC 12228).
Antimicrobial susceptibility
The isolates were tested for susceptibility to oxacillin, vancomycin and teicoplanin by the agar disk diffusion method with Mueller-Hinton agar plates (Difco, USA) according to Clinical Laboratory and Standards Institute (CLSI 2005) recommendations and confirmed by Etest (AB Biodisk, Sweden). The susceptibility to novobiocin, polymyxin B, bacitracin, desferrioxamine and fosfomycin was determined according to Monsen et al. (10).
PFGE typing
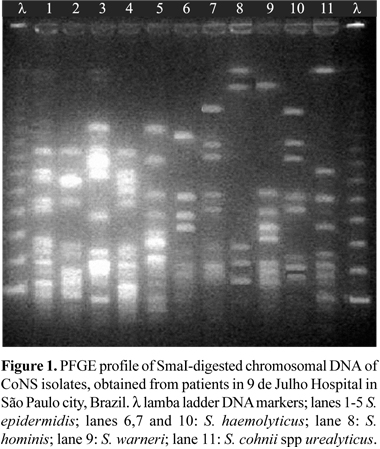
Chromosomal DNA CoNS was prepared in agarose blocks and digested with SmaI (New England BioLabs, USA), as described elsewhere (14). The isolates were run on a 1% agarose gel (Invitrogen, USA) in a CHEF DRIII system (Bio-Rad, USA) under the following conditions: run time, 23 h; temperature, 13ºC; voltage, 200 V; initial forward time, 5 s; final forward time, 60 s. The molecular weight markers (New England BioLabs, USA) were run in the first and in the last lane. The gels were stained with ethidium bromide, washed in water, and photographed under UV light by using the Gel Doc 1000 system (Bio-Rad, USA). The gel patterns were read by visual inspection. The isolates were classified as identical if they shared the same band profile, and isolates differing by more than six bands were considered to represent distinct DNA types (22).
RESULTS AND DISCUSSION
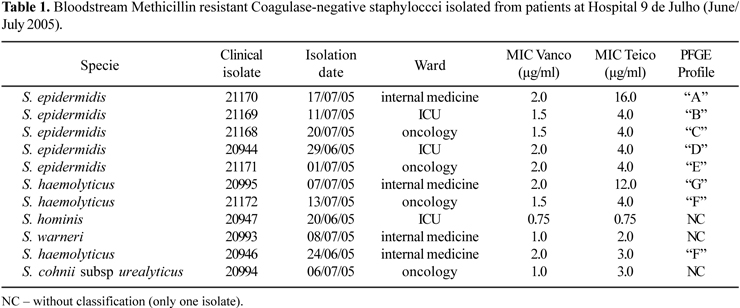
A total of eleven CoNS isolates belonging to five different species were identified including S. epidermidis (5 isolates), S. haemolyticus (3 isolates), S. hominis (1 isolate), S. warneri (1 isolate) and S. cohnii subsp urealyticus (1 isolate) (Table 1). All isolates, except the number 20994, were identified to the species level by the conventional method of Kloos and Banermann (7,8). The isolate 20994 could not be identified by the conventional method, so it was identified by Vitek-2 (bioMèrieux, France) as S. cohnii subsp urealyticus.
The two species most frequently encountered were S. epidermidis and S. haemolyticus.
The S. epidermidis and S. hominis isolates were identified by the disk diffusion susceptibility test to desferrioxamine, since other species of CoNS are resistant to desferrioxamine. To differentiate S. epidermidis from S. hominis, other phenotypic tests were used, as fermentation of trehalose (negative for S. epidermidis), alkaline phosphatase (positive for S. epidermidis) and growth in thioglicolate (positive for S. epidermidis).
The test for production of urease allowed the differentiate S. haemolyticus (urease negative), from S. epidermidis, S. hominis and S. warneri (urease positive). The test of positive PYR along with the hemolytic properties in sheep blood agar and absence of fermentation of mannose allowed the differentiation of S. haemolyticus, the second most prevalent species.
S. cohnii subsp urealyticus (isolate 20994) was the less common specie. Classical tests of resistance to novobiocin, production of urease and absence of sucrose fermentation confirmed the identification of the specie. A discrepancy in the alkaline phosphatase production was noted: the result was negative in the conventional test and positive in the automated system.
All isolates were methicillin-resistant by the disk diffusion test, with MICs > 256 µg/ml by E-Test. Two isolates (S. epidermidis and S. haemolyticus) presented reduced susceptibility to teicoplanin (Table 1). Strains with this characteristic have been reported by Nunes et al. (13) and may be associated with treatment failures or may become precursors of glycopeptide-resistant strains (18).
S. cohnii subsp urealyticus is an unusual opportunist species that has been found in hospital environment like pediatric ICUs (24), and may constitute a dangerous reservoir of multiple antimicrobial plasmid mediated resistance genes (21,23).
Among the five clinical isolates of S. epidermidis five different patterns of PFGE were observed, indicating absence of clonal dissemination among the patients. The same did not occurr with the three clinical isolates of S. haemolyticus, where two isolates from patients at different wards, more than one month apart, presented the same profile, suggesting nosocomial transmission (Fig. 1).
In the last years, the importance of CoNS has been increasing due to their pathogenicity and involvement in human diseases. Their identification species in the clinical laboratories is important but not an easy task, because classical phenotypic tests do not differentiate them from other staphylococci require more time in the identification compared to commercial kits. Many clinical laboratories use automated systems for identification of Staphylococcus spp., although the reliability of results for certain species is not always satisfactory, particularly for species other than S. epidermidis.
Two isolates, one S. epidermidis (isolate 21170) and one S. haemolyticus (isolate 20995) presented high MIC for teicoplanin, but were susceptible to vancomycin (Table 1).
At 9 de Julho hospital, staphylococcal infections, mainly those caused by S. aureus, have been successfully treated with teicoplanin instead of vancomycin, for more than a decade, particulary IV-cateter related infections. In counter part, treatment of central nervous infections and endocarditis with teicoplanin has been less effective, probably due to the occurrence of oxacillin-resistant CoNS also resistant to teicoplanin. A surveillance program of glycopeptide resistance and adequate CoNS specie identification have great importance in determination of risk factors and implementation of nosocomial infection control measures.
ACKNOWLEDGEMENTS
The authors thank Dra Marines Martino from Albert Eistein Microbiology Laboratory for technical support and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação Faculdade Federal de Ciências Médicas de Porto Alegre (FFFCMPA) for financial support.
Submitted: September 15, 2007; Returned to authors for corrections: January 24, 2008; Approved: October 22, 2008
* Corresponding Author. Mailing address: Rua Sarmento Leite 245/204. Cep: 90050-170 Porto Alegre, RS, Brasil. Phone: 55 11 33038740 Fax: 55 11 33038810. E-mail: pedro_dazevedo@yahoo.com.br
- 1. Caierão, J.; Superti, S.; Dias, C.A.G.; d'Azevedo, P.A. (2006). Automated systems in the identification and determination of methicillin resistance among coagulase negative staphylococci. Mem. Inst. Osw. Cruz, 101(3): 277-9.
- 2. Drozenova, J.; Petras, P. (2000). Characteristics of coagulase-negative staphylococci isolated from hemocultures. Epidemiol. Mikrobiol. Imunol., 49(2): 51-8.
- 3. Ferreira, R.B.R.; Nunes, A.P.F.; Kokis, V.M.; Krepsky, N.; Fonseca, L.S.; Bastos, M.C.F.; Marval, M.G.; Santos, K.R.N. (2002). Simultaneous detection of the mec A and ileS-2 genes in coagulase-negative staphylococci isolated from Brazilian hospitals by multiplex PCR. Diagn. Microbiol. Infect. Dis., 42: 205-12.
- 4. Ferreira, R.B.R.; Iorio, N.; Malvar, K.; Nunes, A.; Fonseca, L.; Bastos, C.; Santos, K. (2003). Coagulase-negative Staphylococci: comparison of phenotypic and genotypic oxacilin sucseptibility tests and evaluation of the Agar screening test by using different concentration of oxacilin. J. Clin. Microbiol., 41(8): 3609-14.
- 5. Heikens, E.; Paauw, A.; Florijn, A.; Fluit, A.C. (2005). Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative. Staphylococci. J. Clin. Microbiol., 43(5): 2286-90.
- 6. John, M.A.; Pletch, C.; Hussain, Z. (2002). In vitro activity of quinupristin/dalfopristin, linezolid,telithromicyn and comparator antimicorbial agents against 13 species of coagulse-negative Staphylococci. J. Antimicrob. Chemother., 50: 933-938.
- 7. Kloos, W.E.; Bannerman, T.L. (1994). Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev., 7: 117-140.
- 8. Kloos, W.E.; Bannerman, T.L. (1999). Staphylococcus and Micrococcus.In: Murray, P.R.; Baron, E.J.; Pfaller, M.A.; Tenover, F.C.; Yolken, R.H. (eds), Manual of Clinical Microbiology, 7th ed., ASM Press, Washington DC, 264 pp.
- 9. Marshall, S.A.; Wilke, W.W.; Pfaller, M.A.; Jones, R.N. (1998). Staphylococcus aureus and coagulase-negative staphylococci from blood stream infections: frequency of occurrence, antimicrobial susceptibility, and molecular (mecA) characterization of oxacillin resistance in the SCOPE Program. Diag. Microbiol. Infect. Dis., 30:205-14.
- 10. Monsen, T.; Ronnmark, M.; Olofsson, C.; Wistrom, J. (1998). An inexpensive and reliable method for routin identification of Staphylococcal species. Eur J Clin Microbiol. Infect. Dis., 17: 327-35.
-
11National Committee for Clinical Laboratory Standards NCCLS. (2000). Performance Standards for Antimicrobial Susceptibility Testing, 7th ed. M2-A7. Wayne, Pennsylvania, USA.
-
12National Nosocomial Infections Surveillance System - NNIS. (2003). Am. J. Infect. Control, 31: 481-98.
- 13. Nunes, A.P.F.; Teixeira, L.M.; Iorio, N.L.P.; Bastos, C.C.; Fonseca, L.S.; Souto-Padrón, T.; Santos, K.R.N. (2006). Heterogeneous resistance to vancomycin in Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus warneri clinical strains: characterisation of glycopeptide susceptibility profiles and cell wall thickening. Int. J. Antimicrol. Agents, 27: 307-15.
- 14. Pfaller, M.A.; Hollis, R.J.; Sader, H.S. (1992). Molecular biologyPFGE analysis of chromosomal restriction fragments. In: HD Isenberg (ed). Clinical Microbiology Procedures Handbook, American Society of Microbiology, Washington, D.C., p.10.5.c.1-10.5.c.11.
- 15. Poyart, C.; Quesne, G.; Boumaila, C.; Trieu-Cuot, P. (2001). Rapid and accurate species-level identification of coagulase-negative Staphylococci by using the sod A gene as a target. J. Clin. Microbiol., 39(12): 4296-301.
- 16. Raponi, G.; Ghezzi, M.C.; Gherardi, G.; Dicuonzo, G.; Caputo, D.; Vendittu, M.; Rocco, M.; Micozzi, A.; Mancini, C. (2005). Antimicrobial Susceptibility, Biochemical and Genetic profiles of Staphylococccus haemolyticus strains isolates from the bloodstreams of patients hospitalized in critical care units. J. Chemother., 17(93): 264-9.
- 17. Sader, H.S.; Jones, R.N.; Gales, A.C.; Silva, J.B.; Pignatari, A.C. (2004). SENTRY Antimicrobial Surveillance Program Report: Latin American and Brazilian Results for 1997 trough 2001. Braz. J. Infect. Dis., 8(1): 25-79.
- 18. Sieradzki, K.; Villari, P.; Tomasz, A. (1998). Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin- resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother., 42: 100-07.
- 19. Szarapinska-Kwaszewska, J.; Farkas, L.I. (2003). Synthesis of siderophores by strains of Staphylococcus cohnii isolated from various environments. Acta Microbiol. Pol., 52(3): 261-9.
- 20. Szewczyk, E.M.; Rozalskz, M. (2000). Staphylococcus cohnii-resident of hospital environment: cell-surface features and resistance to antibiotics. Acta Microbiol. Pol., 49(2): 121-33.
- 21. Szewczyk, E.M.; Rozalskz, M.; Cieslikowski, T.; Nowak, T. (2004). Plasmids of Staphylococcus cohnii isolated from the intensive-care unit. Folia Microbiol, 49(2):123-31.
- 22. Tenover, F.C.; Arbeit, R.D.; Goering, R.V. (1995). Interpreting chromosomal DNA restriction patterns produced by Pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol., 33: 2233-9.
- 23. Waldon, E.; Szewczyk, E.M. (2002). Ability of Staphylococcus cohnii strains to adhere to epithelial cells and solid surfaces in the hospital environment. Med. Dosw. Mikrobiol, 54(2): 109-18.
- 24. Yamashita, S.; Yonemura, K.; Sumimoto, R.; Tokunaga, M.; Uchino, M. (2005). Staphylococcus cohnii as a cause of multiple brain abscesses in Weber-Christian disease. J. Neurol. Sci., 15(1-2): 97-100.
Publication Dates
-
Publication in this collection
06 Apr 2009 -
Date of issue
Dec 2008
History
-
Received
15 Sept 2007 -
Accepted
24 Jan 2008