Abstracts
The extended-spectrum β-lactamase (ESBL)-producing bacteria have been isolated at increasing frequency worldwide. Expression of ESBL is often associated with multidrug resistance and dissemination by resistance plasmids. During a two-month period in 2000, 133 clinical isolates of enterobacterial strains were randomly collected from outpatients and inpatients at a university hospital in Turkey. The ESBL producing strains were determined by double-disk synergy (DDS) testing. Twenty ESBL producing strains (15%) including Escherichia coli (n = 9), Klebsiella pneumoniae (n = 7), Klebsiella oxytoca (n = 2) and Enterobacter aerogenes (n = 2) were detected and further analyzed for their resistance transfer features, plasmid profile and nature of the resistance genes. Plasmid transfer assays were performed using broth mating techniques. TEM- and SHV- genes were analyzed by polymerase chain reaction (PCR) and hybridization using specific probes. EcoRI restriction enzyme analyses of R plasmids were used in the detection of epidemic plasmids. Fourteen plasmid profiles (A, B1, B2, C1, and C2 to L) were obtained with EcoRI restriction enzyme analysis. Most of these plasmids were detected to carry both TEM- and SHV-derived genes by PCR, and confirmed by localizing each gene by hybridization assay. Epidemiological evidence indicated that there was an apparent horizontal dissemination of conjugative R plasmids among multidrug-resistant enterobacterial genera and species in this hospital
horizontal spread; resistance plasmids; ESBL; transferência horizontal; ESBL; plasmídios de resistência
O isolamento de bactérias produtoras de beta-lactamases de espectro expandido (ESBL) está aumentando no mundo todo. Freqüentemente, a expressão de ESBL está associada com resistência a múltiplas drogas e disseminação por plasmídios de resistência. Durante um período de dois meses em 2000, 133 isolados clínicos de cepas de enterobactérias foram obtidos aleatoriamente de pacientes internos e externos de um hospital universitário na Turquia. As cepas produtoras de ESBL foram identificadas pelo teste de sinergia em disco-duplo (DDS). Foram detectadas vinte cepas produtoras de ESBL, entre as quais Escherichia coli (n=9), Klebsiella pneumoniae (n=7), Klebsiella oxytoca (n=2) e Enterobacter aerogenes (n=2), que foram posteriormente analisadas quanto a suas características de transferência de resistência, perfil plasmidial e natureza dos genes de resistência. Os testes de transferência de plasmídios foram realizados empregando técnicas de conjugação em caldo. Os genes TEM e SHV foram analisados pela reação da polimerase em cadeia (PCR) e hibridização com sondas especificas. A detecção de plasmídios epidêmicos foi feita por análise dos plasmídios R com a enzima de restrição EcoRI. Através desta análise, foram obtidos catorze perfis plasmidiais (A, B1, B2, C1 e C2 até L).Observou-se pela PCR que a maioria dos plasmidios carregavam genes derivados de TEM e SHV, confirmados através da detecção dos genes pelos testes de hibridização. As evidencias epidemiológicas indicaram que havia uma aparente transferência horizontal dos plasmídios R conjugativos entre as enterobactérias multiresistentes neste hospital.
MEDICAL MICROBIOLOGY
Horizontol dissemination of TEM- and SHV-typr beta-lactamase genes-carrying resistance plasmids amongst clonical isolates of Enterobacteriaceae
Disseminação horizontal de plasmídios de resistência contendo genes de beta-lactamase dos tipos TEM e SHV entre isolados clínicos de Enterobacteriaceae
Osman Birol OzgumusI,II,* * Corresponding Author. Mailing address: Department of Biology, Faculty of Arts & Sciences. Rize University, 53100 Rize, Turkey. Tel.: +90 464 2235375 / ext. 1220. Fax: +90 464 2235376. E-mail: microbsman@yahoo.com ; Ilknur TosunII; Faruk AydinII; Ali Osman KilicI,** ** Current Address: Air Liquide, 200 GBC Drive, Newark, DE 19702. USA
IDepartment of Biology, Faculty of Arts and Sciences, Rize University, 53100 Rize
IIDepartment of Microbiology and Clinical Microbiology, Faculty of Medicine, Karadeniz Technical University, 61080 Trabzon, Turkey
ABSTRACT
The extended-spectrum β-lactamase (ESBL)-producing bacteria have been isolated at increasing frequency worldwide. Expression of ESBL is often associated with multidrug resistance and dissemination by resistance plasmids. During a two-month period in 2000, 133 clinical isolates of enterobacterial strains were randomly collected from outpatients and inpatients at a university hospital in Turkey. The ESBL producing strains were determined by double-disk synergy (DDS) testing. Twenty ESBL producing strains (15%) including Escherichia coli (n = 9), Klebsiella pneumoniae (n = 7), Klebsiella oxytoca (n = 2) and Enterobacter aerogenes (n = 2) were detected and further analyzed for their resistance transfer features, plasmid profile and nature of the resistance genes. Plasmid transfer assays were performed using broth mating techniques. TEM- and SHV- genes were analyzed by polymerase chain reaction (PCR) and hybridization using specific probes. EcoRI restriction enzyme analyses of R plasmids were used in the detection of epidemic plasmids. Fourteen plasmid profiles (A, B1, B2, C1, and C2 to L) were obtained with EcoRI restriction enzyme analysis. Most of these plasmids were detected to carry both TEM- and SHV-derived genes by PCR, and confirmed by localizing each gene by hybridization assay. Epidemiological evidence indicated that there was an apparent horizontal dissemination of conjugative R plasmids among multidrug-resistant enterobacterial genera and species in this hospital.
Key-words:Enterobacteriaceae, horizontal spread, resistance plasmids, ESBL.
RESUMO
O isolamento de bactérias produtoras de beta-lactamases de espectro expandido (ESBL) está aumentando no mundo todo. Freqüentemente, a expressão de ESBL está associada com resistência a múltiplas drogas e disseminação por plasmídios de resistência. Durante um período de dois meses em 2000, 133 isolados clínicos de cepas de enterobactérias foram obtidos aleatoriamente de pacientes internos e externos de um hospital universitário na Turquia. As cepas produtoras de ESBL foram identificadas pelo teste de sinergia em disco-duplo (DDS). Foram detectadas vinte cepas produtoras de ESBL, entre as quais Escherichia coli (n=9), Klebsiella pneumoniae (n=7), Klebsiella oxytoca (n=2) e Enterobacter aerogenes (n=2), que foram posteriormente analisadas quanto a suas características de transferência de resistência, perfil plasmidial e natureza dos genes de resistência. Os testes de transferência de plasmídios foram realizados empregando técnicas de conjugação em caldo. Os genes TEM e SHV foram analisados pela reação da polimerase em cadeia (PCR) e hibridização com sondas especificas. A detecção de plasmídios epidêmicos foi feita por análise dos plasmídios R com a enzima de restrição EcoRI. Através desta análise, foram obtidos catorze perfis plasmidiais (A, B1, B2, C1 e C2 até L).Observou-se pela PCR que a maioria dos plasmidios carregavam genes derivados de TEM e SHV, confirmados através da detecção dos genes pelos testes de hibridização. As evidencias epidemiológicas indicaram que havia uma aparente transferência horizontal dos plasmídios R conjugativos entre as enterobactérias multiresistentes neste hospital.
Palavras-chave:Enterobacteriaceae, transferência horizontal, ESBL, plasmídios de resistência.
INTRODUCTION
Beta-lactam agents including penicillins, cephalosporins, monobactams and carbapenems are among the most frequently prescribed antibiotics worldwide (23). Bacterial resistance to β-lactam antibiotics is increasing throughout the world, mainly through the spread of plasmid-encoded extended-spectrum β-lactamases (ESBLs) (4). ESBLs were first recovered in Klebsiella pneumoniae and have spread to different genera of Enterobacteriaceae, which cause serious therapeutic problems in most developed and developing countries (17). Increasing rates of ESBL-producing Enterobacteriaceae isolates from either the community or the hospital setting have been reported (26).
After oxyimino-β-lactams came into clinical use, resistance to these agents through ESBLs appeared in K. pneumoniae, Escherichia coli strains and other genera. These pathogens have been shown to transfer this resistance to other bacteria via multi-resistant plasmids (16). Therefore, the ESBL genes are usually located on large conjugative plasmids, often carrying genes conferring resistance to aminoglycosides (19). Some are located within transposable elements which strongly facilitate their spread between DNA replicons, even between bacterial strains of different species (4,15). Resistance plasmids are transferred between bacterial strains by conjugation mechanism. The ability of resistance plasmids to transfer among different bacterial species by conjugation is of clinical importance, since they contribute to increased spread of antibiotic resistance in hospitalized patients (9).
Some authors have reported major single-strain outbreaks (12,24), whereas others emphasized the importance of plasmid transfer rather than the strain spread (3). The current study demonstrates that the R plasmids transferring and spreading the multidrug-resistance between different enterobacterial genera and species isolated from both hospitalized patients and outpatients in a 600-bed university hospital.
MATERIAL AND METHODS
Patients and bacterial strains
One hundred and thirty-three enterobacterial Gram negative clinical isolates were randomly recovered from the various clinical specimens (including urine, tracheal aspirate, blood or cerebrospinal fluid) of the patients suffering from various infectious diseases. The samples obtained from the patients who admitted mainly to pediatrics and, intensive care units, surgical services, and the outpatient policlinics during a two-month interval from June to July 2000 at the 600-bed teaching hospital of Karadeniz Technical University, Trabzon, Turkey. Of 133 enterobacterial isolates, ESBL producing 20 strains, E. coli (n = 9), K. pneumoniae (n = 7), K. oxytoca (n = 2) and Enterobacter aerogenes (n = 2) were selected for further epidemiological study. All strains were identified at the species level using the Sceptor System (Becton-Dickinson Microbiology Systems, Sparks, MD, USA). The ESBL-producing clinical isolates were used as donors and E. coli K-12 strain J53-2 (Fmet pro Rifr) as recipient in conjugation experiments. Plasmid DNA purified from E. coli strain V517 was used as plasmid marker (21). E. coli ATCC 25922 was used as control in susceptibility testing. E. coli 7604 and E. coli J53-2 strains served as TEM-1 and SHV-3 type of β-lactamase (carried on plasmid pUD18) positive controls, respectively, for double-disk synergy (DDS) testing and PCR assays.
Antimicrobial susceptibility testing
The susceptibility tests of the clinical strains were carried out by the Sceptor System (Becton-Dickinson Microbiology Systems, Sparks, MD, USA). Results were expressed as resistant (R), intermediate (I) or susceptible (S). Susceptibility of the transconjugants to the antibiotics was determined by standard disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (6). The following antibiotic disks (Oxoid, England) were used in susceptibility testing of the transconjugants: amoxicillin/clavulanate (10 µg/10 µg), cefazolin (30 µg), cefuroxime (30 µg), ceftazidime (30 µg), cefotaxime (30 µg); ceftriaxone (30 µg), cefepime (30 µg), aztreonam (30 µg), gentamicin (10 µg), tetracycline (30 µg) and trimethoprim-sulfamethoxazole (1.25 µg/23.75 µg).
Clinical isolates and the transconjugants were screened for ESBL production according to the CLSI criteria (6), and confirmed by the double-disk synergy (DDS) tests on Mueller-Hinton agar (Difco, USA) as described by Jarlier and colleagues (18).
Transfer of resistance and plasmid analysis
Conjugation assays were performed by broth mating method. Briefly, donor (the ESBL-producing clinical isolates) and recipient (E. coli J53-2) cells grown with agitation in Luria Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl, pH 7.4) were mixed and incubated for 18 h at 35ºC. Transconjugants were selected on Methylene Blue agar (Oxoid, England) supplemented with 150 µg/mL of rifampin (Hoecst, Germany) and 100 µg/mL of ampicillin (Fisher Scientific, USA). The frequency of transfer was expressed relative to the number of donor cells.
Plasmid DNA was purified from clinical isolates and E. coli transconjugants by alkaline lysis method as previously described by Manniatis et al. (22), and digested with EcoR1 restriction endonuclease (MBI Fermentas, USA). Digested DNA fragments were separated by agarose gel electrophoresis, stained with 0.5µg/mL of ethidium bromide (Sigma, USA) and visualized with ultraviolet light. The approximate sizes of the large plasmids were estimated by comparison to plasmid marker (E. coli V517) (21) and confirmed by adding up the restriction fragments.
PCR, Southern blotting and Hybridization assay
The TEM and SHV type β-lactamase genes were amplified using the intragenic oligonucleotide primers as previously described by Arlet and Philippon (1). The primers used for blaTEM were; OT1 (5'-TTGGGTGCACGAGTGGGTTA-3') and OT2 (5'-TAATTGTTGCCGGGAAGCTA-3'), to amplify a 504-bp fragment, and for blaSHV genes, primers OS1 (5'-TCGGG CCGCGTAGGCATGAT-3' and OS2 (5'-AGCAGGGCGAC AATCCCGCG-3') were used to amplify a 626-bp fragment. The PCR products were analyzed by agarose (2%) gel electrophoresis and visualized under ultraviolet light.
Two sets of agarose gels containing EcoRI-digested plasmid DNA fragments were transferred onto two separate nylon membranes (27). The TEM- and SHV-specific probes were generated with PCR using the intergenic primers and plasmid templates harboring blaTEM (pUC18) and blaSHV (pUD18) genes, respectively. The probes were separately labeled with digoxygenin as described by the manufacturer (Bohringer-Mannheim, Germany) and hybridized under high stringency conditions at 68ºC.
RESULTS
Antimicrobial susceptibility of isolates
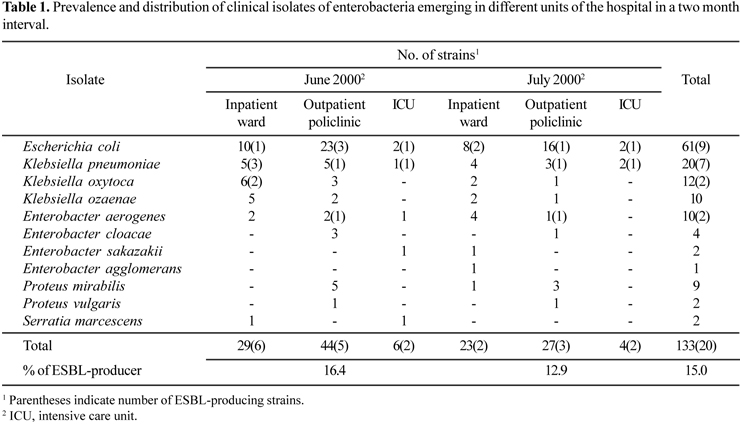
Prevalence and distribution of clinical isolates of enterobacteria emerging in different hospital units are shown in Table 1. Escherichia coli was the most commonly isolated pathogen followed by Klebsiella spp. and Enterobacter spp. Antimicrobial susceptibility profiles and β-lactamase types of ESBL-producing isolates are summarized in Table 2. The susceptibility results indicated that nearly all isolates were resistant to piperacillin but susceptible to imipenem, cefotetan and amikacin. Seven isolates were resistant to nitrofurantoin whereas all isolates except for Enterobacter aerogenes strain TRE164 were susceptible to ciprofloxacin. The only effective antimicrobial among the non-β-lactam antibiotics to all strains seems to be an aminoglycoside antibiotic, amikacin.
ESBL production and plasmid transfer
Twenty of the 133 isolates, which represents 15% of the enterobacterial strains, produced ESBL and showed a typical positive synergy test between cefotaxime, ceftazidime, aztreonam or cefepime and clavulanic acid, but were susceptible to imipenem and cefotetan, consistent with an ESBL production. The ESBL-gene carrying strains represented with a ratio of 16.4% and 12.9% in the months June and July, respectively (Table 1). All ESBL-producing strains including 10 E. coli, 6 K. pneumoniae, 2 K. oxytoca and 2 E. aerogenes were able to transfer their ESBL genes to a recipient E. coli strain J53-2 by conjugation at a transfer frequency of 5x10-8 to 10-4. Co-transfer of gentamicin, tetracycline or trimethoprim/sulfamethoxazole resistance occurred in three cases (Table 3). Most of the transconjugants showed similar antibiotic resistance profile as donors.
Isolates carrying ESBL determinants were obtained most frequently from urine samples in both hospital and community patients, six inpatients and seven outpatients (Table 2). The remaining samples were obtained from respiratory tract infections (four inpatients), bloodstream infections (one inpatient), and cerebrospinal infections (one inpatient). Among the hospitalized patients, half of the ESBL-producing isolates were detected in pediatric-associated wards (Table 2).
PCR detection of blaTEMand blaSHV genes, plasmid profile and hybridization
Multiple β-lactamase genes were identified in 20 strains by TEM- and SHV-specific PCR. Five strains contained single β-lactamase gene, blaSHV. Twelve strains harbored both blaTEM and blaSHV genes whereas three strains had neither blaTEM nor blaSHV genes (Table 2). The sizes of the plasmids recovered from the transconjugants ranged from about 7 to over 80 kb (Fig. 1 and Table 3).
The conjugative plasmids showed 14 different EcoRI restriction enzyme profiles (A, B1, B2, C1, C2, D, E, F, G, H, I, J, K, and L) (Fig. 2A). The first seven profiles (A to E) were harbored by more than one strain, confirming them to be epidemic plasmids, and the others (F to L) were determined as nonepidemic as they depicted different digestion patterns and sporadic emergence.
Two K. pneumoniae isolates, TRE025 and TRE037, were cultured from tracheal aspirate from two unrelated patients. One of the samples was recovered from a patient who suffered from a respiratory infection in pediatrics-intensive care unit (P-ICU) in July, while the other one was from the urine sample of a patient with urinary tract infection (UTI) in a pediatric surgical ward in June. EcoRI restriction analyses showed that these strains harbored two very similar plasmids (Fig 1, profile A).
E. aerogenes TRE088 and E. coli TRE090 strains were isolated from urine specimens of two unrelated outpatients, both with UTI, in June. Both strains harbored two very similar plasmids (Fig. 1, profile B2). Similarly, K. oxytoca TRE063 and E. aerogenes TRE164 strains were isolated from urine samples of a patient with UTI in the pediatric surgical ward in June, and from tracheal aspirate of another patient with respiratory infection in the infectious diseases ward in July, respectively. They carried profile B1 plasmids. The EcoRI restriction pattern of profile B1 plasmid consisted of eight fragments. An additional fragment was observed in B2 profile which indicates the presence of the additional small plasmid (Fig. 1 and Fig. 2A). E. coli TRE153 and K. pneumoniae TRE044 strains were isolated from tracheal aspirate of a patient with lung infection in P-ICU in June, and from urine of another patient with UTI in the infant ward in July, respectively. These two different bacteria harbored a large conjugative plasmid with about 87 kb in size (Fig. 1, profile C2). Surprisingly, K. pneumoniae TRE165 isolated from tracheal aspirate of a patient with respiratory infection from P-ICU carried a small plasmid in addition to a large plasmid of about 87 kb (Fig. 1, profile C1). Plasmid profile C1 consists of seven fragments, one of which belongs to the additional small plasmid (Fig. 1 and Fig. 2A). Remaining plasmid profiles (F to L) were determined as nonepidemic plasmids due to different restriction fragment patterns.
The presence of TEM- or SHV-derived β-lactamase genes was also confirmed by hybridization of EcoRI restriction digested conjugative plasmids with blaTEM-1 (Fig. 2B) and blaSHV-3-specific probes (Fig. 2C). Both probes hybridized to both epidemic and non-epidemic plasmids digested with EcoRI enzyme.
DISCUSSION
Extended-spectrum β-lactamase-producing organisms can be introduced into ICUs. Epidemics of infection from ICUs to other parts of the hospital have been well documented (13). It has been reported that most enterobacteria are responsible for the dissemination of multidrug resistance plasmids in the hospital environment (11). Plasmid-encoded ESBLs often contain resistance determinants for other classes of antimicrobial agents and are readily transmissible from strain-to-strain and between different species of enteric Gram-negative bacilli (8,12). During two months, ESBL-producing enterobacterial genera were mainly isolated from different patients hospitalized in the P-ICU, infant ward and from the pediatrics outpatient policlinics. The patients hospitalized in pediatrics-associated wards were at increased risk of infection because of young age and prolonged period of hospitalization involving various invasive therapeutic and diagnostic procedures. Such factors have been reported to contribute to infection caused by ESBL-producing bacteria (23). Several epidemics have been detected among neonates, elderly patients and even outpatients (2).
The same plasmid in E. coli, K. oxytoca or E. aerogenes strains of unrelated origin is an important finding in regards to the possibility of horizontal spread of the resistance plasmid from one unit to entire hospital. Some ESBL outbreaks have been attributed to the dissemination of plasmids among strains of the family Enterobacteriaceae (19). In other case, spread of a given ESBL in a single environment has been reported due to the occurrence of the same gene within unrelated plasmids (30). Although plasmid dissemination has been well documented (2,9), the spread of an epidemic strain remains the most commonly reported mechanism of ESBL dissemination (13). In this study, detection of very similar, if not identical, R plasmids carried by different enterobacterial genera, isolated from different patients in different times, indicates that the plasmid transfer is quite frequent in the hospital environment.
Discovery of novel transposons encoding an ESBL on a self-transmissible plasmid in a hospital where large quantities of extended-spectrum β-lactam antibiotics are used raises concern for the increased spread of resistance to these groups of antibiotics (15). According to the hospital records regarding β-lactam use in a five month interval in the year 2000 (involving the period of present study), the amount of amoxicillin/clavulanate, ceftazidime, cefotaxime, ceftriaxone, cefepime and aztreonam uses were ca. 0.234, 0.065, 0.008, 0.036, 0.239, 0.053 and 0.002 kg/day/all wards, respectively (data not shown). It is likely that excessive use of antimicrobials in the healthcare environment resulted in the selective increase of R plasmid bearing isolates belonging to different genera of Enterobacteriaceae. It has been reported that constant selective pressure exerted by β-lactams is a risk factor for selection of resistant ESBL-producing strains (14). Similarly, it was also confirmed that genetically different strains could acquire the same plasmid(s), either randomly or specifically due to antibiotic therapy, thus leading to plasmid epidemics (25).
TEM- or SHV-gene carrying isolates were detected by PCR and confirmed the genes localizing on the conjugative plasmids by hybridization assays. Detection of ESBL genes should be confirmed only by sequencing of the PCR products because the genes detected by PCR or hybridization could be the non-ESBL variants TEM-1, TEM-2 or SHV-1. SHV β-lactamase genes are actually present in all K. pneumoniae strains, but once they were transferred by conjugation, and the transconjugant isolates were resistant to extended spectrum β-lactam antibiotics, these β-lactamases should be indeed ESBLs. However, all families of β-lactamases should be probed for the epidemiological importance of β-lactamase genes in Europe, especially the OXA (7) and PER (29) family of enzymes that have been prevalent at other sites in Turkey. Recently, Tasli and Bahar (28) reported that 52.7% TEM, 74.3% SHV, and 32.4% TEM and SHV genes in clinical isolates of Enterobacteriaceae were determined by PCR from western Turkey, and TEM-1, SHV-2, SHV-5 and SHV-12 type enzymes were found to be predominant.
The suitable β-lactam choice seems to be imipenem in treatment of serious infectious diseases caused by enterobacteria in our hospital. However, it is known that the heavy use of carbapenems may favor the selection of Stenotrophomonas maltophilia which is naturally resistant to these drugs (20). Moreover, epidemiological typing of this nonfermenting Gram-negative organism revealed that three small outbreaks occurred from June 2000 to December 2001 in this hospital (5).
The presence of a plasmid-mediated resistance to extended-spectrum β-lactams and to aminoglycosides and capable of intra- and inter-generic spread among enterobacteria are the most important part of our experience in this hospital.
In conclusion, the results of the epidemiological analysis reported here reflect that the multidrug-resistant bacteria can continuously prompt to disseminate antibiotic resistance genes to other clinical pathogens under the constant selective pressure arising from the excess β-lactam usage in the hospital unless effective and strict precautions are taken. Antibiotic policies that restrict the use of cephalosporins, as well as the aminoglycosides, even though the strains were susceptible to amikacin, may be required in addition to infection control policies in this hospital.
ACKNOWLEDGEMENTS
We thank Dr. George A. Jacoby (Lahey Hitchcock Clinic, Department of Infectious Diseases, Burlington, Massachussets, USA) for providing us E. coli J53-2 (pUD18) and E. coli 7604. We also thank Dr. Metin Otkun (Trakya University, Faculty of Medicine, Department of Microbiology and Clinical Microbiology, Edirne, Turkey) for kindly providing us with rifampicin resistant E. coli K-12 strain J53-2.
This study was supported by Karadeniz Technical University Research Foundation (KTU 20.114.001.10).
Submitted: September 20, 2007; Returned to authors for corrections: October 31, 2007; Approved: October 22, 2008
- 1. Arlet, G.; Philippon, A. (1991). Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB). FEMS Microbiol. Lett., 66, 19-25.
- 2. Asensio, A.; Oliver, A.; Gonzáles-Diego, P. et al (2000). Outbreak of a multiresistant Klebsiella pneumoniae strain in an intensive care unit: antibiotic use as risk factor for colonization and infection. Clin. Infect. Dis., 30, 55-60.
- 3. Bingen, E.H.; Desjardins, P.; Arlet, G.; Bourgeois, F.; Mariani-Kurkdjian, P.; Lambert-Zechovsky, N.Y.; Denamur, E.; Philippon, A.; Elion, J. (1993). Molecular epidemiology of plasmid spread among extended broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. J. Clin. Microbiol., 31, 179-184.
- 4. Bush, K.; Jacoby, G.A.; Medeiros, A.A. (1995). A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother., 39, 1211-1233.
- 5. Caylan, R.; Kaklikkaya, N.; Aydin, K.; Aydin, F.; Yilmaz, G.; Ozgumus, B.; Koksal, I. (2004). An epidemiological analysis of Stenotrophomonas maltophilia strains in a university hospital. Jpn. J. Infect. Dis., 57:37-40.
-
6Clinical and Laboratory Standards Institute (CLSI). (2003). Performance standards for antimicrobial disk susceptibility tests. 8th ed. Approved standard, M2-A8, Wayne, Pa (USA).
- 7. Danel, F.; Hall, L.M.; Gur, D.; Livermore, D.M. (1998). OXA-16, a further extended-spectrum variant of OXA-10 β-lactamase, from two Pseudomonas aeruginosa isolates. Antimicrob. Agents Chemother., 42, 3117-3122.
- 8. De Freitas, A.L.P.; Machado, D.P.; Soares, F.S.C.; Barth, A.L. (2003). Extended-spectrum β-lactamases in Klebsiella spp. and Escherichia coli obtained in a Brazilian teaching hospital: detection, prevalence and molecular typing. Braz. J. Microbiol., 34, 344-348.
- 9. Essack, S.Y.; Hall, L.M.; Pillay, D.G.; McFadyen, M.L.; Livermore, D.M. (2001). Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum β-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother., 45, 88-95.
- 10. Fierer, J.; Guiney, D. (1999). Extended-Spectrum β-lactamases. JAMA, 281, 561-564.
- 11. Fosse, T.; Savey, A.; Buisson-Touati, C.; Fabry, J. (2000). Prospective survey of multi-resistant bacteria in 1998: a multicenter study in south-east of France. Infect. Cont. Hosp. Epidemiol., 21, 130.
- 12. French, G.; Shannon, K.P.; Simmons, N. (1996). Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and β-lactam-β-lactamase inhibitor combinations by hyperproduction of SHV-5 β-lactamase. J. Clin. Microbiol., 34, 358363.
- 13. Hart, C.A.; Percival, A. (1982). Resistance to cephalosporins amongst gentamicin-resistant Klebsiellae. J. Antimicrob. Chemother., 9, 275-286.
- 14. Helfand, M.S.; Bonomo, R.A. (2005). Current challenges in antimicrobial chemotherapy: the impact of extended-spectrum-β-lactamases and metallo-β-lactamases on the treatment of resistant gram-negative pathogens. Curr. Opin. Pharmacol., 5, 452-458.
- 15. Heritage, J.; Hawkey, P.M.; Todd, N.; Lewis, I.J. (1992). Transposition of the gene encoding a TEM-12 extended-spectrum β-lactamase. Antimicrob. Agents Chemother., 36, 1981-1986.
- 16. Jacoby, G.A. (1997). Extended-spectrum β-lactamases and other enzymes providing resistance to oxyimino-β-lactams. Infect. Dis. Clin. North. Am., 11, 875-886.
- 17. Jacoby, G.A.; Medeiros, A.A. (1991). More extended-spectrum β-lactamases. Antimicrob. Agents Chemother., 35, 1697-1704.
- 18. Jarlier, V.; Nicolas, M.H.; Fournier, G.; Philippon, A. (1988). Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis., 10, 867-878.
- 19. Legakis, N.J.; Tzouvelekis, L.S.; Hatzoudis, G.; Tzelepi, E.; Gourkou, A.; Pitt, T.L.; Vatopoulos, A.C. (1995). Klebsiella pneumoniae infections in Greek hospitals. J. Hosp. Infect., 31, 177-87.
- 20. Luzzaro, F.; Mantengoli, E.; Perilli, M. et al (2001). Dynamics of a nosocomial outbreak of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum β-lactamase. J. Clin. Microbiol., 39, 1865-1870.
- 21. Macrina, F.L.; Kopecko, D.J.; Jones, K.R.; Ayers, D.J.; McCowen, S.M. (1978). A multiple plasmid containing Escherichia coli strain: convention source of size reference plasmid molecules. Plasmid, 1, 417-420.
- 22. Manniatis, T.; Fristch, E.F.; Sambrook, J. (1982). Molecular cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- 23. Medeiros, A.A. (1997). Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis., (Suppl. 1), S19-S45.
- 24. Otman, J.; Perugini, M.E.; Tognim, M.C.B.; Vidotto, M.C. (2007). Atypical phenotypic characteristics of klebsiella pneumoniae isolates from an outbreak in a neonatal intensive care unit in Brazil. Braz. J. Microbiol., 38, 273-277.
- 25. Petit, A.; Gerbaud, G.; Sirot, D.; Courvalin, P.; Sirot, J. (1990). Molecular epidemiology of TEM-3 (CTX-1) β-lactamase. Antimicrob. Agents Chemother., 34, 219-224.
- 26. Pitout, J.D.; Nordmann, P.; Laupland, K.B.; Poirel, L. (2005). Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother., 56, 52-59.
- 27. Southern, E.M. (1975). Detection of specific sequences among DNA fragments separated by agarose gel electrophoresis. J. Mol. Biol., 98, 503-517.
- 28. Tasli, H.; Bahar, I.H. (2005). Molecular characterization of TEM- and SHV-derived extended-spectrum β-lactamases in hospital-based Enterobacteriaceae in Turkey. Jpn. J. Infect. Dis., 58, 162-167.
- 29. Vahaboglu, H.; Ozturk, R.; Aygun, G.; Coskunkan, F.; Yaman, A.; Kaygusuz, A.; Leblebicioglu, H.; Balik, I.; Aydin, K.; Otkun, M. (1997). Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nation-wide multi-centre study. Antimicrob. Agents Chemother., 41, 2265-2269.
- 30. Wiener, J.; Quinn, J.P.; Bradford, P.A.; Goering, R.V.; Nathan, C.; Bush, K.; Weinstein, R.A. (1999). Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA, 281, 517-523.
Publication Dates
-
Publication in this collection
06 Apr 2009 -
Date of issue
Dec 2008
History
-
Accepted
31 Oct 2007 -
Received
20 Sept 2007