Abstracts
PtSRR1 EST was previously identified in the first hours of Pisolithus tinctorius and Castanea sativa interaction. QRT-PCR confirmed PtSRR1 early expression and in silico preliminary translated peptide analysis indicated a strong probability that PtSRR1 be a transmembrane protein. These data stimulate the PtSRR1 gene research during ectomycorrhiza formation.
ectomycorrhiza; symbiosis related genes/proteins; Pisolithus tinctorius
PtSRR1 foi isolado preliminarmente de P. tinctorius nas primeiras horas da interação com raízes de C. sativa. Análises de QRT-PCR confirmaram sua expressão positiva (12 h) e seu peptídeo putativo indicou forte possibilidade para proteína transmembranar. Estes dados estimulam o estudo do PtSRR1 durante a formação de ectomicorrizas.
ectomicorriza; genes/proteínas simbiose- regulados; Pisolithus tinctorius
ENVIRONMETAL MICROBIOLOGY
PtSRR1, a putative Pisolithus tinctorius symbiosis related receptor gene is expressed during the first hours of mycorrhizal interaction with Castanea sativa roots
PtSRR1, um possível receptor simbiose-regulado de Pisolithus tinctorius é expresso nas primeiras horas de interação ectomicorrízica com raízes de Castanea sativa
B. Acioli-SantosI,II,III; E. MalossoI; C. E. Calzavara-SilvaII; C. E. P. LimaI; A. FigueiredoIII; M. SebastianaIII; M. S. PaisIII
IDepartamento de Micologia, Universidade Federal de Pernambuco, Recife, PE, Brasil
IICentro de Pesquisas Aggeu Magalhães, Fundação Instituto Oswaldo Cruz, Departamento de Virologia e Terapia Experimental-LaViTE, Universidade Federal de Pernambuco, Recife, PE, Brasil
IIIUnit of Molecular Biology and Plant Biotechnology, ICAT, Faculdade de Ciências de Lisboa, Universidade de Lisboa, Lisboa, Portugal
Corresponding Author Corresponding Author B. Acioli-Santos Departamento de Micologia, Universidade Federal de Pernambuco Av. Prof. Nelson Chaves s/n, Cidade Universitária 50670-420, Recife, PE, Brasil Phone: +55 81 2126 8865. Fax: +55 81 21268482 E-mail: bartacioli@cpqam.fiocruz.br
ABSTRACT
PtSRR1 EST was previously identified in the first hours of Pisolithus tinctorius and Castanea sativa interaction. QRT-PCR confirmed PtSRR1 early expression and in silico preliminary translated peptide analysis indicated a strong probability that PtSRR1 be a transmembrane protein. These data stimulate the PtSRR1 gene research during ectomycorrhiza formation.
Key-words: ectomycorrhiza, symbiosis related genes/proteins, Pisolithus tinctorius.
RESUMO
PtSRR1 foi isolado preliminarmente de P. tinctorius nas primeiras horas da interação com raízes de C. sativa. Análises de QRT-PCR confirmaram sua expressão positiva (12 h) e seu peptídeo putativo indicou forte possibilidade para proteína transmembranar. Estes dados estimulam o estudo do PtSRR1 durante a formação de ectomicorrizas.
Palavras-chave: ectomicorriza, genes/proteínas simbiose- regulados, Pisolithus tinctorius.
The formation of ectomycorrhiza is a process governed by a complex biochemical and molecular interaction between the two partners before physical contact. Several stages of the ectomycorrhiza formation and maintenance processes from preinfection to the formation of the mantle and the Hartig net have been described, and it is obvious that changes in gene expression have to accompany the processes leading to symbiosis (6,8).
Studies evaluating the fungal transcript pattern during symbiosis formation have demonstrated that mycorrhization induces changes in the expression of genes normally expressed in the free organisms, without the participation of symbiosis specific genes (9). In this paper, we present a fungal cDNA EST representing a gene that is upregulated at 12 h of interaction between P. tinctorius and C. sativa (1). Its expression, the putative protein structure and its possible function in the symbiosis are discussed.
Biological material acquisition/maintenance and ectomicorrhizal induction is described by Baptista et al. (2007) (2). Micohhiza stimulated ("myc") and control mycelium (only in water) were harvested 12 h after contact, snap-frozen in liquid nitrogen and stored at -80ºC. A cDNA library of P. tinctorius was constructed from 6 µg of mRNA mix (control and "myc") using the SMART cDNA Library Construction Kit (BD Clontech, Palo Alto, CA, U.S.A) as presented by Acioli-Santos et al. (2008). For the quantification of the PtSRR1 mRNAs, the reverse transcription of each target RNA (control RNA and "myc" at 6 h and 12 h of interaction) was carried out (7).
The cloned PtSRR1 EST fragment is 432 bp long. An untranslated region is observed downstream from the putative open reading frame (Fig. 1). The PtSRR1 sequence has 70% similarity to a sequence of Pisolithus microcarpus (CB010071), a fungus that forms ectomycorrhiza with Eucalyptus. The putative PtSRR1 peptide has 48% similarity to a protein of the fungus Schizophyllum commune (AF335537) that is upregulated under low nitrogen conditions. The study of the PtSRR1 expression using QRT-PCR allowed the confirmation of the up-regulation at 12 h of interaction, revealing positive transcription rates 1350 fold higher than the control. At 6 h of fungus-plant interaction, the relative values were close to one, suggesting that changes in the transcription levels may occur between 6 and 12 h of interaction.
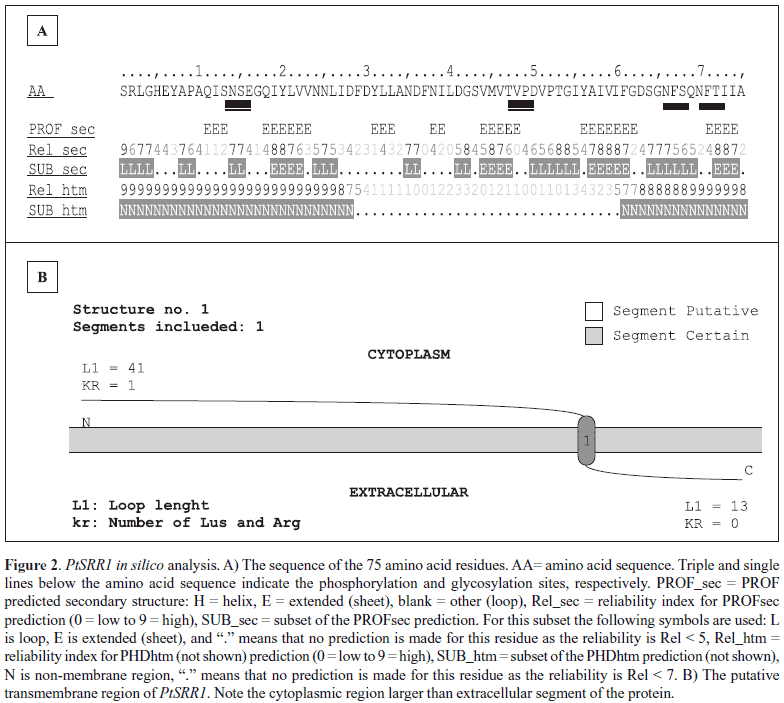
In silico translation of PtSRR1 nucleotide sequence resulted in a peptide fragment of 75 amino acids (8.2 kDa), without the initial methionine. No cysteine residues were found in the PtSRR1 amino acid sequence. The analyses of the PtSRR1 peptide primary structure (http://ca.expasy.org/cgi-bin/prosite) enabled the identification of four post-translational modification sites as follows: two N-glycosylation sites with high probability of occurrence between the residues 66 to 69 (NFSQ) and 70 to 73 (NFTI), and two Casein Kinase II phosphorylation sites, in the positions 13 to 16 (SNSE) and 47 to 50 (TVPD), respectively (Fig. 2a). No usual protein domais were identified. Secondary PtSRR1 structure analisys showed abundance of beta-structures (Fig. 2a). No helix was detected. The peptide shows a well-defined transmembrane region, despite the low probability suggested by its analysis (http://www.predictprotein.org). It was not possible to obtain a PtSRR1 three-dimensional model based on homology modeling (http://www.swissmodel.expasy.org/ SWISS-MODEL.html) (Fig. 2b).
The expression of several genes at 6 h of interaction between Laccaria bicolor and Pinus resinosa has been reported (4,5). However, most of the differentially expressed fungal genes were observed in later stages of symbiotic development, especially after two or more days of interaction (3,6,9), which is corroborated by the 12 h PtSRR1 transcription. Therefore, the high relative expression of PtSRR1 at 12 h favours its investigation. QRT-PCR data confirmed the cDNA microarrays analysis of the fungal PtSRR1 and its high relative transcription at 12 h of ectomycorrhizal interaction. Transcription of this gene does not occur until 6 h of contact, suggesting that this period between 6 and 12 h can be critical for its expression.
The PtSRR1 gene is that probably triggered by the low availability of nitrogen that could function as an "indicator" of host root proximity. PtSRR1 homologue peptide (AF335537) was identified in Schizophyllum commune. This homologue peptide presents high expression when the mycelium is growing under low nitrogen availability conditions. Further physiological studies and the acquisition of the complete ORF of this gene are necessary for functionality tests in the symbiosis. These results would allow to understand the real function of the PtSRR1 protein.
As the PtSRR1 amino acid sequence is not complete and the three-dimensional protein structure is not known, any conclusion about the role of this protein is premature. However, considering its secondary structure prediction, the PtSRR1 seems to be a transmembrane protein with an intracellular segment containing at least one phosphorylation accessible site and an extracellular region containing two glycosylation sites. Thus, there is a possibility that the PtSRR1 acts as membrane receptor/extra-intracellular signal-transducer element through sites of glycosylation and phosphorylation, or be a secreted protein. However, in silico data obtained using the truncated PtSRR1 amino acid sequence would differ from the full-length amino acid sequence. These data strongly stimulate the research of PtSRR1 gene role in the ectomycorrhizal process as a potential marker/regulator of the early stages of symbiotic interaction.
ACKNOWLEDGEMENTS
This work was partially supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior/MCT-Brazil) as a Ph.D. scholarship and grant to the first author.
Submitted: May 20, 2008; Returned to authors for corrections: September 27, 2008; Approved: March 31, 2009
- 1. Acioli-Santos, B.; Sebastiana, M.; Pessoa, F.; Sousa L.; Figueiredo, A.;Fortes, A.M.; Baldé, A.; Maia, L.C.; Pais, M.S. (2008). Fungal transcript pattern during the preinfection stage (12 h) of ectomycorrhiza formed between pisolithus tinctorius and castanea sativa roots, identified using cdna microarrays. Curr. Microbiol (In press)
- 2. Baptista, P.; Martins, A.; Pais, M.S.; Tavares, R.M.; Lino-Neto, T. (2007). Involvement of reactive oxygen species during early stages of ectomycorrhiza establishment between Castanea sativa and Pisolithus tinctorius Mycorrhiza, 17, 185-193.
- 3. Duplessis, S.; Courty, P.; Tagu, D.; Martin, F. (2005).Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus New Phytol., 165, 599-611.
- 4. Kim, S.; Bernreuther, D.; Thumm, M.; Podila, G. (1999). LB-AUT7, a novel symbisis-regulated gene from an ectomycorrhizal fungus, Laccaria bicolor, is functionally related to vesicular transport and autophagocytosis. J. Bacteriol., 181, 1963-1967.
- 5. Kim, S.; Zheng, J.; Hiremath, T.; Podila, G.K. (1998). Cloning and characterization of a symbiosis-related gene from an ectomycorrhizal fungus Laccaria bicolor Gene, 222, 203-212.
- 6. Le Quéré, A.; Wright, D.P.; Söderström, B.; Tunlid, A.; Johansson, T. (2005). Global patterns of gene regulation associated with the development of ectomycorrhiza between Birch (Betula pendula Roth.) and Paxillus involutus Mol. Plant-Microbe Interact., 18, 659-673.
- 7. Santos, B.A. (2006). Expressão gênica no fungo Pisolithus tinctorius em etapas iniciais da interação ectomicorrízica com Castanea sativa: estudo do padrão de transcrição e de possíveis fatores de regulação da simbiose. Tese de Doutorado Lisboa, 169 p. (Ph. D., Departamento de Biologia Vegetal, Universidade de Lisboa).
- 8. Tagu, D.; Lapeyrie, F.; Martin, F. (2002). The ectomycorrhizal symbiosis: genetics and development. Plant Soil, 244: 97-105.
- 9. Voiblet, C.; Duplessis, S.; Encelot, L.; Martin, F. (2001). Identifications of symbiosis-reguleted genes in Eucalyptus globulus-Pisolitus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant. J, 25, 181-191.
Publication Dates
-
Publication in this collection
24 July 2009 -
Date of issue
June 2009
History
-
Accepted
31 Mar 2009 -
Reviewed
27 Sept 2008 -
Received
20 May 2008