Abstracts
The Alternaria brown spot (ABS) is a disease caused in tangerine plants and its hybrids by the fungus Alternaria alternata f. sp. citri which has been found in Brazil since 2001. Due to the recent occurrence in Brazilian orchards, the epidemiology and genetic variability of this pathogen is still an issue to be addressed. Here it is presented a survey about the genetic variability of this fungus by the characterization of twenty four pathogenic isolates of A. alternata f. sp. citri from citrus plants and four endophytic isolates from mango (one Alternaria tenuissima and three Alternaria arborescens). The application of two molecular markers Random Amplified Polymorphic DNA (RAPD) and Amplified Fragment Length Polymorphism (AFLP) had revealed the isolates clustering in distinct groups when fingerprintings were analyzed by Principal Components Analysis (PCA). Despite the better assessment of the genetic variability through the AFLP, significant modifications in clusters components were not observed, and only slight shifts in the positioning of isolates LRS 39/3 and 25M were observed in PCA plots. Furthermore, in both analyses, only the isolates from lemon plants revealed to be clustered, differently from the absence of clustering for other hosts or plant tissues. Summarizing, both RAPD and AFLP analyses were both efficient to detect the genetic variability within the population of the pathogenic fungus Alternaria spp., supplying information on the genetic variability of this species as a basis for further studies aiming the disease control.
Alternaria brown spot; molecular markers; citrus plants; PCA analysis; genetic variability
A mancha marrom ou mancha de Alternaria é uma doença causada pelo fungo Alternaria alternata f. sp. citri, encontrada no Brasil desde 2001 em plantas de tangerina e seus híbridos. Por se tratar de uma doença recente no Brasil, a epidemiologia e variabilidade genética deste patógeno compõem importantes pontos a serem estudados. Este trabalho teve como objetivo avaliar a variabilidade genética deste patógeno por meio da caracterização de vinte e quatro isolados de A. alternata f. sp. citri de plantas de citros juntamente com quatro isolados endofíticos de manga, sendo um deles identificado como Alternaria tenuissima e outros três como Alternaria arborescens. A análise de componentes principais (PCA) do perfil de bandas obtidos pela aplicação de duas técnicas de marcadores moleculares, Amplificação Aleatória de Polimorfismos de DNA (RAPD) e Polimorfismo de Comprimento de Fragmentos Amplificados (AFLP), mostrou a formação de quatro grupos distintos. Apesar do mais amplo perfil de análise por meio da técnica de AFLP, não foi observado nenhuma modificação significativa dentro dos grandes grupos obtidos quando comparado ao RAPD, exceto no posicionamento dos isolados LRS 39/3 e 25M. Além disso, em ambas as análises, somente os isolados de plantas de limão agruparam entre si. Considerando outros hospedeiros ou tecidos de planta não foi possível encontrar grupos específicos. Concluindo, ambas as análises (RAPD e AFLP) são eficientes no estudo de variabilidade genética de Alternaria spp., fornecendo informações sobre a diversidade genética desta espécie, servindo como base para futuramente correlacionar este estudo com estudos adicionais objetivando o controle da doença.
Mancha marrom; marcadores moleculares; citrus; análise de PCA; variabilidade genética
ENVIRONMENTAL MICROBIOLOGY
Genetic variability of Brazilian isolates of Alternaria alternata detected by AFLP and RAPD techniques
Variabilidade genética de isolados Brasileiros de Alternaria alternata por meio de marcadores moleculares de AFLP e RAPD
Francisco Dini-AndreoteI; Vivian Cristina PietrobonI; Fernando Dini AndreoteI; Aline Silva RomãoI; Marcel Bellato SpósitoII; Welington Luiz Araújo I, III,* * Corresponding Author. Mailing address: Núcleo Integrado de Biotecnologia Universidade de Mogi das Cruzes, Av. Dr. Cândido Xavier de Almeida Souza, 200, 08780-911 Mogi das Cruzes, SP, Brasil..; Fone: (55) 11 4798 7000 Ramal 7340, (55) 11 4798 7106.; E-mail: welingtonluiz@umc.br and wlaraujo@esalq.usp.br
IDepartamento de Genética, Escola Superior de Agricultura "Luiz de Queiroz", Universidade de São Paulo, Piracicaba, SP, Brasil
IIDepartamento Científico, Fundo de Defesa da Citricultura, Araraquara, SP, Brasil
IIINúcleo Integrado em Biotecnologia, Universidade de Mogi das Cruzes, Mogi das Cruzes, SP, Brasil
ABSTRACT
The Alternaria brown spot (ABS) is a disease caused in tangerine plants and its hybrids by the fungus Alternaria alternata f. sp. citri which has been found in Brazil since 2001. Due to the recent occurrence in Brazilian orchards, the epidemiology and genetic variability of this pathogen is still an issue to be addressed. Here it is presented a survey about the genetic variability of this fungus by the characterization of twenty four pathogenic isolates of A. alternata f. sp. citri from citrus plants and four endophytic isolates from mango (one Alternaria tenuissima and three Alternaria arborescens). The application of two molecular markers Random Amplified Polymorphic DNA (RAPD) and Amplified Fragment Length Polymorphism (AFLP) had revealed the isolates clustering in distinct groups when fingerprintings were analyzed by Principal Components Analysis (PCA). Despite the better assessment of the genetic variability through the AFLP, significant modifications in clusters components were not observed, and only slight shifts in the positioning of isolates LRS 39/3 and 25M were observed in PCA plots. Furthermore, in both analyses, only the isolates from lemon plants revealed to be clustered, differently from the absence of clustering for other hosts or plant tissues. Summarizing, both RAPD and AFLP analyses were both efficient to detect the genetic variability within the population of the pathogenic fungus Alternaria spp., supplying information on the genetic variability of this species as a basis for further studies aiming the disease control.
Key words:Alternaria brown spot, molecular markers, citrus plants, PCA analysis, genetic variability.
RESUMO
A mancha marrom ou mancha de Alternaria é uma doença causada pelo fungo Alternaria alternata f. sp. citri, encontrada no Brasil desde 2001 em plantas de tangerina e seus híbridos. Por se tratar de uma doença recente no Brasil, a epidemiologia e variabilidade genética deste patógeno compõem importantes pontos a serem estudados. Este trabalho teve como objetivo avaliar a variabilidade genética deste patógeno por meio da caracterização de vinte e quatro isolados de A. alternata f. sp. citri de plantas de citros juntamente com quatro isolados endofíticos de manga, sendo um deles identificado como Alternaria tenuissima e outros três como Alternaria arborescens. A análise de componentes principais (PCA) do perfil de bandas obtidos pela aplicação de duas técnicas de marcadores moleculares, Amplificação Aleatória de Polimorfismos de DNA (RAPD) e Polimorfismo de Comprimento de Fragmentos Amplificados (AFLP), mostrou a formação de quatro grupos distintos. Apesar do mais amplo perfil de análise por meio da técnica de AFLP, não foi observado nenhuma modificação significativa dentro dos grandes grupos obtidos quando comparado ao RAPD, exceto no posicionamento dos isolados LRS 39/3 e 25M. Além disso, em ambas as análises, somente os isolados de plantas de limão agruparam entre si. Considerando outros hospedeiros ou tecidos de planta não foi possível encontrar grupos específicos. Concluindo, ambas as análises (RAPD e AFLP) são eficientes no estudo de variabilidade genética de Alternaria spp., fornecendo informações sobre a diversidade genética desta espécie, servindo como base para futuramente correlacionar este estudo com estudos adicionais objetivando o controle da doença.
Palavras-chave: Mancha marrom, marcadores moleculares, citrus, análise de PCA, variabilidade genética.
INTRODUCTION
The disease Alternaria brown spot (ABS) was first described in 1903 in Australia (10), where it was found infecting tangerines and their hybrids, in leaves, twigs, and immature fruit, causing decrease in yield and commercial value of fruits (17). Besides the fruit damages, this disease can also cause intense leaf fall, drought of branches and premature fruit drop from plants. In fruits the injuries are brown with a depression in the center, often showing liquid exudation with a viscose consistency surrounded by a yellow halo. The center of the injuries may become leathery. In branches, injuries are necrotic, variable in size and with brown color, while in leaves the symptoms are characterized by brown or black small injuries with yellow halos around the spot.
The genetic variability of this pathogen is scarcely explored, constituting an important subject to be evaluated due to the persistence of this disease in the citrus orchards. Molecular techniques based in DNA markers, such as the random amplified polymorphic DNA (RAPD) (2,23) and the amplified fragment length polymorphism (AFLP) (18,21), are very informative and costeffective approaches for assessing genetic diversity of a wide range of fungi, such as Agaricus blazei (6), Alternaria species (1,3,9), Erythricium salmonicolor (19), Lactobacillus (20), Metharizium anisopliae (8), and Trypanosoma (13). The main advantage of such techniques over the onegene based assessment is the genomewide biodiversity which does not require prior availability of sequences on databases and are therefore readily applicable to any organism. However, while RAPD relies on the use of a single primer for PCR at low stringency, in a simple, fast and little DNA amount requiring, the AFLP is a DNA profiling technique based on the amplification of restriction fragments by PCR and further detection of a large numbers of informative bands on the DNA polymorphisms. The AFLP reliability results from the combination of both intrinsic features: the restriction enzyme digestion and the robustness of high stringency PCR (13).
In the present work, it is presented data in order to supply information about the genetic variability found in the population of pathogenic A. alternata and related species, based on the application of two techniques, RAPD and AFLP. Additionally to the genetic variability of the targeted population, the efficacy and the drawbacks of each technique are discussed.
MATERIAL AND METHODS
Brazilian fungal isolates cultivation and DNA extraction
A total of twenty-eight Alternaria spp. isolates were collected from different plant tissues (leafs, fruits and branches), collected from cultivated tangerine and lemons in orchards located in three Brazilian states: Minas Gerais (MG), São Paulo (SP) and Rio Grande do Sul (RS) (Table 1), and kindly supplied by Dr. Eduardo Feichtenberger (Instituto Biológico, Unidade de Pesquisa e Desenvolvimento de Sorocaba, Sorocaba, SP - Brasil). In addition, four leaf endophytic isolates from mango, named 12M, 14M, 16M and 25M, were included in the analysis. All isolates were grown on potato dextrose (PD) medium during seven days at 28ºC and total DNA was extracted with Wizard Genomic DNA Purification kit (Promega, USA).
Characterisation of isolates by RAPD
RAPD analysis was carried out on 25 µL PCR reactions, containing 10ng of template DNA, 0.28 mM of each dNTP, 3.2 mM of MgCl2, 2U of Taq DNA polymerase and 0.45 µM of primers. Reactions were separetely prepared for amplification with primers OPA16 (5'-AGCCAGCGAA-3'), OPC08 (5'-TGGACCGGTG-3'), OPC06 (5'-GAACGGACTC-3'), G13 (5'-CTCTCCGCCA-3'), OPP16 (5'-CCAAGCTGCC-3'), OPP19 (5'-GGGAAGGACA-3'), OPX12 (5'-TCGCCAGCCA-3') and OPX14 (5'-ACAGGTGCTG-3') (Operon Technologies, Brazil). The amplifications were carried out as follows: 5 min at 94ºC, followed by 40 cycles of 1 min at 92ºC, 1 min at 35ºC, 2 min at 72ºC with a final extension at 72ºC for 5 min. DNA fragments were separeted in 1.5% agarose gels, which were stained with ethidium bromide (0.5mg/mL).
Characterisation of isolates by AFLP
AFLP analysis was performed as previously described by Masiga and Turner (14). Briefly, Alternaria spp. genomic DNA (approximately 200 ng) was digested with 5U of EcoRI and MseI endonucleases by incubation overnight at 37 ºC in a 50 µl reaction volume. The reactions were then heat-inactivated at 70ºC for 10min and adapters for EcoRI and MseI were linked to the fragments at 23 ºC for 3h using the T4 DNA ligase. After ligation, two rounds of PCR amplification were performed. The first round was carried out with primers specific for the adapters, EcoRI (core primer with one adenine as selective nucleotide) and MseI (core primer with one adenine/cytosine as selective nucleotides).
Amplification was performed with 26 cycles by denaturation at 94 ºC for 1min, annealing at 56 ºC for 1min, and extension at 72 ºC for 1min. PCR products were then diluted five-fold in TE buffer (10mM Tris-Cl, pH 8.0, 0.1mM EDTA) and used for selective amplification with a specific primer to the EcoRI adapters selective nucleotides (E-A) and to the MseI adapters plus two selective nucleotides (M-CA and M-AT). Selective PCR amplification was carried out as follows: two cycles were performed at 94 ºC for 30 s, 65 ºC for 30 s, and 72 ºC for 1min. The same conditions for denaturation and extension were maintained for 12 cycles, while the annealing temperature was down by to 56 ºC and this was followed by 23 cycles, denaturing at 94 ºC for 30 s, annealing at 56 ºC for 30 s while the extension step was at 72 ºC for 2min.
The visualization of patterns of AFLP generated by each isolate, amplification reactions were mixed with an equal volume (8 µl) of formamide dye (98% formamide, 10mM EDTA, pH 8.0, with bromophenol blue and xylene cyanoll as tracking dyes), heat-inactivated for 5min at 95 ºC and chilled on ice. Fifteen microlitres of each sample was then loaded on a 6% polyacrylamide gel in 1x TBE buffer with a sensor using 80W per gel and maximum 50ºC. After the electrophoresis, the AFLP gel was stained with AgNO3 following the protocol described in Creste et al. (7).
Gel image analyses
The eight gels of RAPD and the two gels of AFLP were visually analyzed, where the patterns of bands were identified in different isolates constituting the fingerprints observed. Tables containing the band positions and the presence in different isolates were used for principal components analyses (PCA) using Canoco (Canoco 4.5, Biometris, Wageningen, the Netherlands).
RESULTS
Random Amplified Polymorphic DNA
Several primers were prior tested for amplification, number of bands and informative polymorphism detection. From the initially testes primers, eight were considered in RAPD analysis, revealing to be reproducible indicating a high-quality of the fungal DNA used as template combined with the standardized RAPD procedure due to the MgCl2 constant concentration. A total of 85 bands ranging from 0.2 to 3.0 kb were observed, revealing to qualitatively varying among the isolates fingerprints. Out of all 85 bands, 71 were polymorphic among the isolates fingerprints, while the remaining 14 bands revealed to be monomorphic. Regarding the polymorphic bands, higher numbers of informative bands were found in analysis with primers C06 and X12.
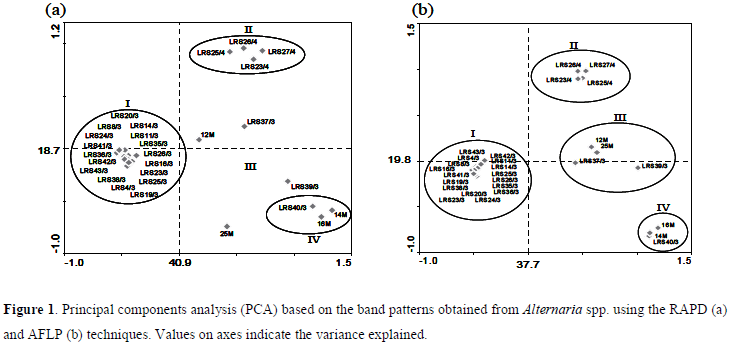
Considering the genetic variability of analyzed isolates, revealed by the clustering of samples by RAPD, observed in the PCA analysis (Fig. 1a), four groups could be observed. However, the formation of groups hardly showed correlation with the host plants from which isolates were obtained. The only exception was the clustering observed for four isolates (cluster II: LRS 23/4, LRS 25/4, LRS 26/4 and LRS 27/4) obtained from lemon plants. Other three observed groups were the cluster I, the biggest group composed by all isolates from tangor "Murcott" and other additional isolates (LRS 04/03, LRS 25/03, LRS 35/03, LRS 36/03, LRS 38/03, LRS 43/03). The cluster IV showed to be constituted by two endophytic isolates from mango (14M and 16M) and one pathogenic isolate from tangerine "Cravo". At last, the cluster III was not clearly formed in RAPD analysis, with samples LRS 37/3, LRS 39/3, 12M and 25M spread in the biplot.
Amplified Fragment Length Poplymorphism
The AFLP analysis using two sets of primers resulted in a total of 151 bands, where 121 were polymorphic among genotypes and the remaining 30 bands were monomorphic, demonstrating the efficacy of this technique in generating higher numbers of informative fingerprints when compared to RAPD.
AFLP fingerprints revealed the formation of IV groups, where correlations with isolates origins were not observed, as previous described in RAPD analysis. Also, similarly to RAPD results were observed, since the clusters observed for the AFLP analysis were maintained except for the better clustering of the cluster III, composed by the isolates 12M, 25M, LRS 37/7 and LRS 39/3 (group III) (Fig. 1b).
Comparative analysis of RAPD and AFLP fingerprinting
In general it is possible to state that both techniques were efficient in detecting the genetic variability of the Alternaria spp. isolates used in the present study. Comparing RAPD and AFLP techniques, although there was a difference in the number of obtained bands, the percentage of polymorphic bands were very similar, 80.13 and 83.52% in AFLP and RAPD analysis, respectively.
Data from RAPD and AFLP analyses were in accordance, with the exception of a better clustering of group III obtained by AFLP. In this study, the efficiency of RAPD and AFLP was confirmed in PCA analysis, which explained 59.6 and 57.5% of the variance for RAPD and AFLP, respectively (Fig.1). In both analyses, the main separation, observed in the x axis, was relative to cluster I from the others clusters II, III and IV. These other clusters were separated on y axis, revealing to be responsive to a secondary component of variance. This secondary separation was more evidenced in AFLP analysis, where the cluster was more consistent for the group III, what was spread in the plotting based on RAPD analysis (Fig. 1).
DISCUSSION
The Brazilian citrus culture presents many phytossanitary problems, including more than 50 fungal diseases (19). In most of these cases, genetic pathogen variability studies are the first step aiming to obtain more information on the disease etiology, following by studies about control disease.
Claes et al. (5) and Masiga et al. (13) reported the lower efficiency of RAPD when compared to AFLP in detecting the genetic variability, mainly regarding the reproducibility of results. However, in the present study, Alternaria spp. isolates were subjected to two fingerprint typing techniques, revealing that both methodologies were efficient in detecting the genetic variability of the analyzed twenty eight isolates, contrasting with previous studies (5,13).
Despite the advantages and disadvantages of both RAPD and AFLP techniques, the present work has revealed the common absence of patterns of Alternaria spp. isolates according to the host plant. The only host which revealed to be selective for certain genotype of this fungus species was the lemon plant. This could corroborate with data where the description of pathogenic Alternaria sp. for lemon plants are classified as Alternaria citri (11). However, previous study shown that in Florida (USA) that population of Alternaria spp. causing brown spot in grapefruit and hybrids (tangerine X grapefruit) may be distinguished by RAPD markers (16). On the other hand geographical populations of Alternaria alternata from California cannot be grouped by RAPD (15), suggesting that the host plant, but not the geographical origin, may play an important role in genetic diversity of this pathogen.
Concerning the lack of groups formation for other host plants, it is also remarkable the non-clustering of the isolates 16M, 14M and 25M, all identified as Alternaria arborescens according to the ITS sequences (data not shown). Similar results were reported by Lopandic et al. (12), which suggest that methods, such as RAPD and AFLP are more discriminative than the ITS sequences. Hence, this could be explained by the fact that dominant markers (RAPD and AFLP) represent specific DNA markers randomly distributed throughout the whole genome (22). On the other hand, the identification by ITS sequence is based in a specific fragment into genomic DNA hosting a conserved region. Moreover, it is important to consider that several factors may affect the estimative of genetic relationships among isolates, such as the number of markers used, the distribution of the markers in the genome of analyzed samples and the nature of the evolutionary mechanisms underlying the variation measured (4). Also, studies including infection analysis should be done to confirm the pathogenicity of LRS37/3, LRS39/3 and LRS40/3, which could be endophytic Altenaria isolated from lesion caused by pathogenic A. alternata.
In conclusion, this study showed two efficient molecular markers to study genetic variability from Alternaria spp., supplying information about the genetic variability of this pathogen. Moreover the understandings of basic factors correlated to the etiology were determined. All these information might serve as a basis for further studies aiming the development of better mechanisms of disease control in Brazilian orchards.
ACKNOWLEDGEMENTS
This work was supported by a grant from FUNDECITRUS (Araraquara, São Paulo State, Brazil) and FAPESP (Foundation for Research Assistance of São Paulo State, Brazil). We thank FAPESP for the fellowship to W.L.A. (Proc. nº 03/10527-4). We also thank Sr. José Antônio da Silva for technical assistance in isolation and identification of mango isolates.
Submitted: August 04, 2008; Returned to authors for corrections: November 03, 2008; Approved: May 03, 2009.
- 1. Aradhya, M.K.; Chan, H.M.; Parfitt, D.E. (2001). Genetic variability in the pistachio late blight fungus, Alternaria alternata. Mycol. Res 105 (3): 300-306.
- 2. Belaj, A.; Satovic, Z.; Ismaili, H.; Panajoti, D.; Rallo, L.; Trujillo, I. (2003). RAPD genetic diversity of Albanian olive germplasm and its relationships with other Mediterranean countries. Euphytica 130 (3), 387-395.
- 3. Bock, C.H.; Thrall, P.H.; Brubaker, C.L.; Burdon, J.J. (2002). Detection of genetic variability in Alternaria brassicicola using AFLP fingerprinting. Mycol. Res 106 (4): 428-434.
- 4. Chowdhury, A.K.; Yonemoto, Y.; Kato, H.; Macha, M.M. (2005). Cultivar identification by morphometric descriptors and RAPD markers among some Acerola (Malpighia glabra Linn.) cultivars. Jpn. J. Trop. Agr 49 (2), 41-42.
- 5. Claes, F.; Agbo, E.C.; Radwanska, M.; Te Pas, M.F.W.; Baltz, T.; De Waal, D.T.; Goddeeris, B.M.; Claassen, E.; Buscher, P. (2003). How does Trypanozoon equiperdum fit into the Trypanozoon group? A cluster analysis by RAPD and multiplex-endonuclease genotyping approach. Parasitol 126 (5), 425-431.
- 6. Colauto, N.B.; Dias, E.S.; Gimenes, M.A.; da Eira, A.F. (2002). Genetic characterization of isolates of the basidiomycetes Agaricus blazei by RAPD. Braz. J. Microbiol. 33: 131-133.
- 7. Creste, S.; Tulmann-Neto, A.; Figueira, A. (2001). Detection of single sequence repeat polymorphism in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol. Biol. Rep 19 (4), 299-306.
- 8. Freire, L.L.C.; da Costa, A.B.L.; Goés, L.B.; de Oliveira, N.T. (2001). DNA polymorphism and total protein in mutants of Metharizium anisopliae var. anisopliae (Metsch.) Sorokin strain E9. Braz. J. Microbiol. 32: 93-97.
- 9. Hong, S.G.; Maccaroni, M.; Figuli, P.J.; Pryor, B.M.; Belisario, A. (2006). Polyphasic classification of Alternaria isolated from hazelnut and walnut fruit in Europe. Mycol. Res 110: 1290-1300.
- 10. Kiely, T.B. (1964). Brown spot of Emperor mandarin. Agric. Gaz. N.S.W. 75 (6), 854-856.
- 11. Laranjeira, F.F.; Amorim, L.; Bergamim-Filho, A.; Aguilar-Vildoso, C.I.; Coletta-Filho, H.D. (2005). Fungos, procariotos e doenças abióticas. In: Mattos-Junior, D., Negri, J.D., Pio, R.M., Pompeu-Junior, J. (eds). Citros Instituto Agronômico e Fundag, Campinas, Brasil, p.511-566.
- 12. Lopandic, K.; Molnár, O.; Prillinger, H. (2005). Application of ITS sequence analysis, RAPD and AFLP fingerprinting in characterising the yeast Genus Fellomyces. Microbiol. Res 160 (1), 13-26.
- 13. Masiga, D.K.; Ndung'u, K.; Tweedie, A.; Tait, A.; Turner, C.M.R. (2006). Trypanosoma evansi: Genetic variability detected using amplified restriction fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) analysis of Kenyan isolates. Exp. Parasitol 114 (3), 147-153.
- 14. Masiga, D.K.; Turner, C.M.R. (2004). Amplified restriction fragment length polymorphism (AFLP) analysis. In: Melville, S.E.(eds). Parasite Genomics Protocols Humana Press, New Jersey, USA, p.173-185.
- 15. Morris, P.F.; Connoly, M.S.; St Clair, D. (2000). Genetic diversity of Alternaria alternate isolated from tomato in California assessed using RAPDs. Mycol. Res 104 (3) 286-292.
- 16. Peever, T.L.; Olsen, L.; Ibañez, A.; Timmer, L.W. (2000). Genetic differentiation and host specificity among populations of Alternaria spp. causing brown spot of grapefruit and tangerine x grapefruit hybrids in Florida. Phytopathology 90: 407-414.
- 17. Reis, R.F.; Almeida, T.F.; Stuchi, E.S.; Goes, A. (2007). Susceptibility of citrus species to Alternaria alternata, the causal agent of the Alternaria brown spot. Sci. Hortic-Amsterdam 113 (4), 336-342.
- 18. Rotondi, A.; Magli, M.; Ricciolini, C.; Baldoni, L. (2003). Morphological and molecular analyses for the characterization of a group of Italian olive cultivars. Euphytica 132 (2), 129-137.
- 19. Sebastianes, F.L.S.; Maki, C.S.; Andreote, F.D.; Araújo, W.L.; Pizzirani-Kleiner, A.A. (2007). Genetic Variability and vegetative compatibility of Erythricium salmonicolor isolates. Sci. Agr 64 (2), 162-168.
- 20. Torriani, S.; Clementi, F.; Vancannety, M.; Hoste, B.; Dellaglio, F.; Kersters, K. (2001). Differentiation of Lactobacillus plantarum, L. pentosus and L. paraplantarum Species by RAPD-PCR and AFLP. Syst. Appl. Microbiol. 24 (8), 554-560.
- 21. Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; Zabeau, M. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acid. Res 23 (21), 4407-4414.
- 22. Wachira, F.; Tanaka, J.; Takeda, Y. (2001). Genetic variation and differentiation in tea (Camellia sinensis) germplasm revealed by RAPD and AFLP variation. J. Hortic. Sci. Biotech 76 (5), 557-563.
- 23. Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acid. Res 18 (22), 6531-6535.
Publication Dates
-
Publication in this collection
18 Aug 2009 -
Date of issue
Sept 2009
History
-
Accepted
03 May 2009 -
Reviewed
03 Nov 2008 -
Received
04 Aug 2008