Abstract
Rapid detection of Mycobacterium tuberculosis complex (MTBC) is a critical step in controlling tuberculosis (TB). In this study, we used IS6110 as the specific identification target to develop a novel hybridization signal amplification method (HSAM) for the rapid and direct detection of MTBC from clinical sputum specimens. This system consists of magnetic bead-linked capture probes for target isolation, dextranbased nanoparticles for amplifying the reporter molecule (biotinylated-FITC), and detection probes (2B-DNA) for binding the nanoparticles. Both the capture and detection probes were specific to the IS6110 target sequence. Our results determined that as few as 10 copies of the IS6110 sequence or 10 M. tuberculosis bacteria could be detected, indicating that the HSAM assay is as sensitive as conventional PCR, and the assay was specific enough to distinguish MTBC from nontuberculosis mycobacteria (NTM). A total of 176 clinical sputum specimens were collected for HSAM evaluation, and the results were compared to those from traditional culture and biochemical identification techniques. This assay had a sensitivity of 88.3%, a specificity of 91.8%, a positive predictive value of 93.8% and a negative predictive value of 84.8% for the detection of MTBC. This technique is highly sensitive and specific, is easy to perform, and does not require any sophisticated detection equipment; thus, this approach has great potential in clinical TB detection and diagnostic applications.
Mycobacterium tuberculosis complex (MTBC); Detection; Diagnosis; Rapid methods; Identification
MEDICAL MICROBIOLOGY
Rapid identification of Mycobacterium tuberculosis complex by a novel hybridization signal amplification method based on self-assembly of DNA-streptavidin nanoparticles
Haihe WangI; Chunyan ZhaoII; Fan LiII,* * Corresponding Author. Mailing address: Department of Pathogenobiology, Norman Bethune College of Medical Sciences, Jilin University, Changchun, China. Fan Li.; Tel: 86-431-85619462.; E-mail: lifan@jlu.edu.cn
IDepartment of Pathogenobiology, Daqing Branch of Harbin Medical University, Daqing, China
IIDepartment of Pathogenobiology, Norman Bethune College of Medical Sciences, Jilin University, Changchun, China
ABSTRACT
Rapid detection of Mycobacterium tuberculosis complex (MTBC) is a critical step in controlling tuberculosis (TB). In this study, we used IS6110 as the specific identification target to develop a novel hybridization signal amplification method (HSAM) for the rapid and direct detection of MTBC from clinical sputum specimens. This system consists of magnetic bead-linked capture probes for target isolation, dextranbased nanoparticles for amplifying the reporter molecule (biotinylated-FITC), and detection probes (2B-DNA) for binding the nanoparticles. Both the capture and detection probes were specific to the IS6110 target sequence. Our results determined that as few as 10 copies of the IS6110 sequence or 10 M. tuberculosis bacteria could be detected, indicating that the HSAM assay is as sensitive as conventional PCR, and the assay was specific enough to distinguish MTBC from nontuberculosis mycobacteria (NTM). A total of 176 clinical sputum specimens were collected for HSAM evaluation, and the results were compared to those from traditional culture and biochemical identification techniques. This assay had a sensitivity of 88.3%, a specificity of 91.8%, a positive predictive value of 93.8% and a negative predictive value of 84.8% for the detection of MTBC. This technique is highly sensitive and specific, is easy to perform, and does not require any sophisticated detection equipment; thus, this approach has great potential in clinical TB detection and diagnostic applications.
Key words:Mycobacterium tuberculosis complex (MTBC); Detection; Diagnosis; Rapid methods; Identification
INTRODUCTION
Tuberculosis (TB) is one of the greatest health concerns in developing and industrialized countries, affecting 1.7 billion people worldwide. There are 8 million new cases and around 3 million deaths annually. Alarmingly, the number of TB cases is rising at a rate of 2% per year. Rapid detection of Mycobacterium tuberculosis complex (MTBC) strains is one of the most important factors in minimizing the spread of this disease. In low- and middle-income countries, conventional smear microscopy and culture are the primary methods for diagnosis of pulmonary tuberculosis. Smear microscopy is easy, fast and inexpensive, but its efficiency is limited by relatively low sensitivity (3, 15, 17, 19). Cultures can take from 6 to 8 weeks to obtain conclusive results, limiting this technique's ability to aid in diagnosis and inform immediate treatment decisions. Recently, several unique detection systems have been developed as rapid tests for the direct identification of MTBC from clinical specimens, including a hybridization assay on microwell plates, a DNA microarray assay and cycling probe technology (2, 5, 14). Variations in sensitivity and the high costs of these tests have prevented the widespread use of these systems for the detection of TB.
We have recently developed a novel hybridization signal amplification method (HSAM) for the rapid identification of MTBC from clinical specimens. In this study, we combine magnetic bead-based DNA isolation, amplification of a reporter molecule (biotinylated-FITC) by nanoparticles and fluorescence detection. IS6110 was selected as a target site for detection (8), and is specifically recognized by the capture and detection probes. The biotinylated capture probe was immobilized on streptavidin-coated Dynal magnetic beads, taking advantage of the high affinity between biotin and streptavidin (STV) (12, 23). The bisbiotinylated detection probe (2B-DNA) was conjugated to DNA-streptavidin nanoparticles (10, 11). Because each nanoparticle provides multiple binding sites for the biotinylated FITC reporter molecule, a thousand-fold amplification can be achieved (Fig. 1). The objective of this study was to determine the sensitivity and specificity of the HSAM assay for MTBC, and to assess its feasibility for the detection of MTBC from clinical specimens.
MATERIALS AND METHODS
Bacterial strains
The Mycobacterium tuberculosis (MTB) H37Rv strain was provided by the Center for Disease Control, China. To produce samples with a known number of bacteria, M. tuberculosis was grown in 7H9 broth medium (4). Several bacterial species were used to evaluate the specificity of the HSAM reaction: Mycobacterium bovis; nontuberculosis mycobacteria (NTMs) including Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium kansasii, Mycobacterium chelonae and Mycobacterium fortuitum; and other bacteria such as Escherichia coli, Staphylococcus aureus, β-hemolytic Streptococcus and Streptococcus pneumoniae.
HSAM assay
The HSAM assay involved several steps. First, the capture probe was immobilized on the magnetic beads in a 100 µl reaction mixture containing 97 µl of binding buffer (20 mM Tris-HCl, 1 M LiCl, 2 mM EDTA), 1 µl of capture probe (2 nM) and 2 µl (105/µl) of Dynabeads® M-280 Streptavidin (Invitrogen Dynal AS, California, USA). The mixture was incubated at 37for 20 min at 200 rpm to allow the streptavidin-coated beads to bind the biotin on the capture probe. Second, the target sequence was hybridized to the capture probe by adding 2 µl of supernatant from a DNA extraction to the reaction mixture and incubating at 55for 1 h. Third, the samples were washed by applying a magnetic field for 5 min to attract the magnetic beads with the attached capture probe/target complex to a specific location in the reaction container. The magnetic beads were washed three times with 400 µl of TE buffer (10 mM Tris-HCl, 1 mM EDTA) at room temperature to remove unhybridized capture probe and target DNA. Fourth, the free biotin-binding sites were blocked by adding 100 µl of blocking buffer (10 mM Tris-HCl, 1 mM EDTA, 800 µM d-Biotin) to the magnetic bead pellet and incubating at 37for 20 min at 200 rpm (11). This process blocked the unoccupied biotin-binding sites of the surface-bound STV on the beads. The samples were then washed three times with 400 µl of TE buffer. Fifth, the nanoparticles were immobilized by adding 95 µl of binding buffer and 5 µl of streptavidin-DNA nanoparticles to the reaction mixture. This mixture was incubated at 55for 1 h to allow the complete hybridization of 2B-DNA to the target. The samples were then washed three times with 400 µl of TE buffer to remove unhybridized nanoparticles. Sixth, the read-out of the HSAM assay was obtained by adding 40 µl of TE buffer and 1 µl (2 µM) of biotinylated FITC to the mixture and incubating the samples at 37for 20 min at 200 rpm. The samples were washed three times with 400 µl of TE buffer to remove unhybridized FITC. Finally, the results of the HSAM assay were obtained using fluorescent microscopy (Olympus Optical Co., Ltd, Tokyo, Japan).
Specimen collection and processing
A total of 176 clinical sputum specimens were collected from patients who presented with clinically and radiologically suspected pulmonary tuberculosis at the Mycobacteriology Laboratory, Hospital of Tuberculosis, Changchun, China between August 2006 and March 2007. The specimens were decontaminated by the addition of an equal volume of sputolysin-NaOH (4%) and were incubated for 20 min at room temperature while rocking. After neutralization with 0.067 M sodium phosphate buffer, pH 5.3 (7, 25), the mixture was centrifuged at 2800×g for 20 min. The supernatant was discarded, and the pellets were resuspended in 1 ml of phosphate buffered saline (PBS, 10 mM Na2HPO4, 2 mM KH2PO4, 0.137 M NaCl, 2.7 mM KCl, pH 7.4).
Culture and biochemical methods for diagnosis of MTBC
About half of each pellet containing suspected pulmonary tuberculosis specimens was inoculated into two Lowenstein-Jensen agar slants and incubated at 37with 5% CO2. Mycobacterial growth was observed over 2-8 weeks after inoculation. Positive cultures for acid-fast bacilli were identified by conventional biochemical tests (8, 16).
Sample preparation for HSAM
To prepare a sample for HSAM evaluation, 500 µl of diluted bacterial culture or sputum pellet was centrifuged at 2800×g for 20 min. The supernatant was removed, and the pellets were resuspended in 500 µl of distilled water. The mixture was vortexed with glass beads and boiled at 100ºC for 10 min (6). This process was repeated three times, and then the mixture was centrifuged at 8000×g for 5 min. The resulting supernatant was used in the HSAM assay.
Probe preparation
The detection (2B-DNA) and capture probes were derived from the nucleic acid sequences encoding the insertion sequence IS6110 (GenBank M29899). The capture probe was derived from nucleic acids at positions 431 to 470 with a biotin label on the 5' end. The detection probe was derived from positions 471 to 530 with the 5' and 3' ends labeled with biotin (TaKaRa Biotechnology (Dalian) Co., Ltd, Dalian, China). IS6110 was selected as a target site that was specifically recognized by the capture and detection probes (Table 1).
Nanoparticle assembly
Dextran-based poly-streptavidin (Poly-STV, 110 µM) (Dako A/S, California, USA) and 2B-DNA were diluted to 5 µM and 1 µM, respectively, with buffer B (1 M Tris-HCl pH 7.5, 0.5 M EDTA, pH 8.0). Nanoparticles were assembled in 30 µl reaction mixtures in the presence of 15 µl Poly-STV (5 µM) and 15 µl 2B-DNA (1 µM). The reaction mixture was incubated at 55 for 30 min at 800 rpm. The products were analyzed by gel electrophoresis using a 1.0% agarose gel and atomic force microscopy.
Validation of HSAM
To evaluate the sensitivity of the test, ten-fold serial dilutions of the synthetic IS6110 DNA target and M. tuberculosis H37Rv were prepared. A culture of M. tuberculosis H37Rv grown in 7H9 broth medium was adjusted to the turbidity equivalent of the No. 4 McFarland barium sulfate nephelometer standard (22). Approximately 105 colony-forming units (CFUs) were contained in 50 µl of the diluted samples. The bacteria were diluted with PBS (pH 7.4) in a series of 10-fold dilutions from 104 to 101bacteria in 2 µl. The IS6110 DNA target was diluted with distilled water from 105 to 101 molecules per 2 µl. M. bovis, five NTM strains, E. coli, β-hemolytic Streptococcus, S. aureus and S. pneumoniae were used to evaluate the specificity of the HSAM assay. The target extraction and HSAM procedures were described above.
Quality control
For the quality control of the HSAM assay, a fixed amount (about 105 CFU/ml) of MTB bacteria was spiked into a normal sputum sample following the same sputum treatment procedures and DNA extraction protocols for clinical sputum samples. For each HSAM assay, positive and negative controls were included.
RESULTS
To establish efficient conditions for nanoparticle formation, molar ratios of 2B-DNA to Poly-STV in the nanoparticle assembly were examined. As shown in Figure 2, a 1:5 ratio produced the maximum amount of nanoparticles that are too large to migrate in a 1% agarose gel, which indicates the formation of nanoparticles (Lane 1). The ratio of 1:1 produced the lowest amount of nanoparticles, and free 2B-DNA is visible (Lane 3). The ratio of 1:2 is intermediate (Lane 2). Atomic force microscopy showed that the self-assembled nanoparticles are three-dimensional ellipsosomes, and the diameter and ramp diameter are 32 nm and 72 nm, respectively. Very few nanoparticles have a diameter of 240 nm, perhaps due to adhesion between the nanoparticles (Fig. 3).
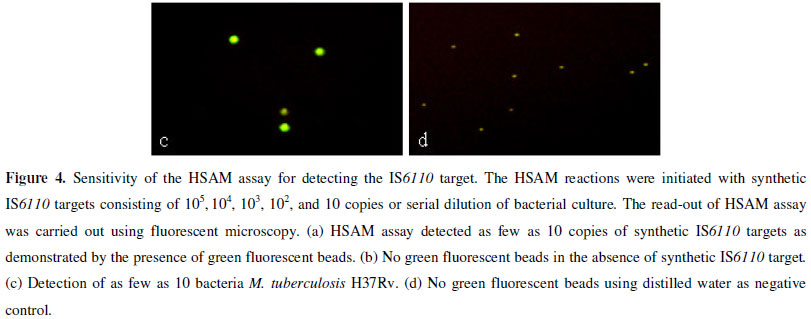
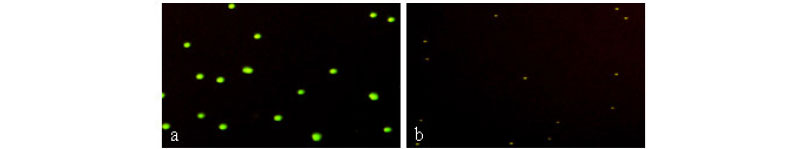
The analytical sensitivity of the HSAM assay was determined by testing serial dilutions of a synthetic IS6110 DNA target and M. tuberculosis H37Rv. We initially determined the sensitivity of the HSAM assay using a ten-fold serial dilution of the synthetic IS6110 DNA target. The lowest detection limit by the HSAM assay was 10 molecules (Fig. 4a), which was confirmed by observing the presence of green fluorescent beads. The assay sensitivity was further analyzed using the M. tuberculosis H37Rv strain. The results in Figure 4c indicate that the HSAM assay was able to detect as few as 10 bacteria.
To determine the assay specificity, we tested the following 11 bacterial strains: M. tuberculosis H37Rv, M. bovis, five NTMs, E. coli, S. aureus, β-hemolytic Streptococcus, and S. pneumoniae. As expected, M. tuberculosis H37Rv and M. bovis were positive for the IS6110 insertion sequence (Fig. 5 a,c), while the five NTMs and other bacteria were negative (Fig. 5 b,d). These results confirmed the specificity of the HSAM assay.
A total of 176 clinical sputum specimens collected during the period August 2006 to March 2007 were diagnosed by traditional culture and biochemical identification methods. In parallel, the spent specimen sediments were subjected to DNA extraction and HSAM analysis, the results of which are summarized in Table 2. A total of 103 specimens were reported to contain MTBC strains through the standard culture and biochemical methods. The HSAM assay identified 97 specimens with MTBC strains, resulting in 88.3% sensitivity, 91.8% specificity, 93.8% positive predictive value and 84.8% negative predictive value. IS6110 was identified in six specimens that were MTBC culture-negative. Among the 103 culture-positive MTBC specimens, 12 were IS6110-negative according to the HSAM assay.
DISCUSSION
Rapid detection of MTBC strains and their differentiation from NTMs are critical for initiating appropriate therapy and facilitating measures to prevent the dissemination of TB. A range of rapid tests based on nucleic acid amplification techniques have been developed for the direct detection of MTBC from clinical samples, including PCR. However, these methods may not satisfy the requirements for common use: high sensitivity and specificity, ease in application and cost-effectiveness (18). The relatively high cost of most molecular methods may be one of the major characteristics that hinder the wide use of these systems, especially in developing countries where TB is more prevalent than in developed ones. The HSAM assay is a novel, simple detection method based on the self-assembly of DNA-STV nanoparticles. This assay is easy to perform, does not require any sophisticated detection equipment and is highly sensitive and specific. In addition, it is fast and inexpensive; it takes less than 5 h from the receipt of specimens to completion of the test.
The HSAM assay is a variation of traditional probe signal amplification schemes based on the concept of combining signal amplification by nanoparticles, magnetic bead-based DNA isolation, and fluorescence detection (13). In this study, we used a synthetic IS6110 DNA target and M. tuberculosis H37Rv to test the HSAM assay. The results obtained by testing ten-fold serial dilutions of the IS6110 target and M. tuberculosis H37Rv indicated an analytical sensitivity of 10 synthetic target molecules or 10 bacteria, respectively. Other researchers have reported a detection range of 1-100 cells for PCR techniques (8, 9, 20, 21). The increased sensitivity of the HSAM assay may be due to several factors. First, the three-dimensional porous nanoparticles provide multiple binding sites for reporter molecules (biotinylated FITC), allowing for a thousand-fold amplification. The self-assembled nanoparticles are composed of bis-biotinylated single-stranded DNA (2B-DNA) and dextran-based poly-streptavidin (Poly-STV). The tetravalent binding capacity of STV, combined with its high affinity for biotin, lead to the formation of the three-dimensional nanoparticles (24). The nanoparticles differ from other DNA-streptavidin nanostructures containing double-stranded DNA and streptavidin that are widely used as reagents in immuno-PCR (10, 11) in which the 2B-DNA is used as a detection probe and is complementary to the DNA target sequence. Second, the HSAM assay detects IS6110, which is present at a level of approximately 10-16 copies per cell, which may enhance the sensitivity of the HSAM assay relative to others that detect target sequences with only a single or very low copy number. Third, Dynal magnetic beads were used as a solid support for the capture probe and as magnetic enrichment for target molecules, allowing the test to be performed directly from processed samples without the need for extensive nucleic acid purification.
While conventional culture and biochemical identification serve as the gold standards for diagnosis, we demonstrated that the HSAM assay is an excellent alternative for the detection of MTBC in 176 clinical sputum specimens. As a positive control, a fixed amount (about 105 CFU/ml) of MTB bacteria was spiked into a normal sputum sample, and distilled water was used as a negative control. Compared with conventional culture and biochemical diagnosis methods, HSAM resulted in 88.3% sensitivity, 91.8% specificity, 93.8% positive predictive value and 84.8% negative predictive value for MTBC diagnosis. The results are comparable to, or even better than, those obtained by PCR for MTBC diagnosis. The sensitivities for PCR range from 74% to 100% (8, 9, 20, 21), and acid fast bacilli (AFB) smear method sensitivities vary from 30% to more than 70% (1). Among the 103 cultures in which MTBC specimens were identified by culture and biochemical techniques, 91 were IS6110-positive in the HSAM assay. In contrast, 12 specimens that were positive by traditional techniques were MTBC-negative in our assay, which may be due to too few bacterial cells in the sputum specimens. However, there were 6 culture-negative, IS6110-positive samples, which could be due to non-specific hybridization during the HSAM assay. The HSAM assay for MTBC detection can be performed anywhere, and there is no need for a contamination-proof hood or specific lab area, as is required for PCR. The HSAM assay can be used to detect different DNA and RNA targets, which is easily accomplished by altering the sequence of the capture and detection probes.
In summary, the present study showed that the HSAM assay can detect MTBC in clinical specimens. The method is highly sensitive and specific, economical, time-saving and simple. Although further adjustments are required to improve its sensitivity and quantitative aspects, we expect that the HSAM assay will be a useful tool for the rapid detection of MTBC.
ACKNOWLEDGMENTS
This study was supported by Department of Pathology, Mount Sinai School of Medicine (New York), and the National Natural Science Foundation of China (30672007).
Submitted: December 23, 2009; Returned to authors for corrections: September 14, 2010; Approved: March 14, 2011.
- 1. Albay, A.; Kisa, O.; Baylan, O.; Doganci, L. (2003). The evaluation of FASTPlaqueTB test for the rapid diagnosis of tuberculosis. Diagn. Microbiol. Infect. Dis. 46(3),211-215.
- 2. Beggs, M.L.; Cave, M.D.; Marlowe, C.; Cloney, L.; Duck, P.; Eisenach, K.D. (1996). Characterization of Mycobacterium tuberculosis complex direct repeat sequence for use in cycling probe reaction. J. Clin. Microbilol 34(12),2985-2989.
- 3. Caws, M.; Wilson, S.M.; Clough, C.; Drobniewski, F. (2000). Role of IS6110-targeted PCR, culture, biochemical, clinical, and immunological criteria for diagnosis of tuberculous meningitis. J. Clin. Microbiol 38(9),3150-3155.
- 4. Flournoy, D.; Twilley, J. (2001). Modified Middlebrook 7H9 broth for the rapid detection of mycobacteria. Clin. Lab. Sci. 14(2),85-88.
- 5. Fukushima, M.; Kakinuma, K.; Hayashi, H.; Nagai, H.; Ito, K.; Kawaguchi, R. (2003). Detection and identification of Mycobacterium species isolated by DNA microarray. J. Clin. Microbiol 41(6),2605-2615.
- 6. Gill, P.; Ramezani, R.; Amiri, M.V.; Ghaemi, A.; Hashempour, T.; Eshraghi, N.; Ghalami, M.; Tehrani, H.A. (2006). Enzyme-linked immunosorbent assay of nucleic acid sequence-based amplification for molecular detection of M.tuberculosis. Biochem. Biophys. Res. Commun 347(4),1151-1157.
- 7. Li, F.; Zhao, C.; Zhang, W.; Cui, S.; Meng, J.; Wu, J.; Zhang, D.Y. (2005). Use of ramification amplification assay for detection of Escherichia coli O157:H7 and other E. coli Shiga toxin-producing strains. J. Clin. Microbiol 43(12),6086-6090.
- 8. Montenegro, S.H.; Gilman, R.H.; Sheen, P.; Cama, R.; Caviedes, L.; Hopper, T.; Chambers, R.; Oberhelman, R.A. (2003). Improved detection of Mycobacterium tuberculosis in Peruvian children by use of a heminested IS6110 polymerase chain reaction assay. Clin. Infect. Dis. 36(1),16-23.
- 9. Marchi, A.M.; Juttel, I.D.; Kawacubo, E.M.; Dalmarco, E.M.; Blatt, S.L.; Cordova, C.M.M. (2008). Evaluation of methods for detection and identification of Mycobacterium species in patients suspected of having pulmonary tuberculosis. Braz. J. Microbiol. 39(4),613-618.
- 10. Niemeyer, C.M.; Adler, M.; Pignataro, B.; Lenhert, S.; Gao, S.; Chi, L.; Fuchs, H.; Blohm, D. (1999). Self-assembly of DNA-streptavidin nanostructures and their use as reagents in immuno-PCR. Nucleic. Acids. Res. 27(23),4553-4561.
- 11. Niemeyer, C.M.; Wacker, R.; Adler, M. (2003). Combination of DNA-directed immobilization and immuno-PCR:very sensitive antigen detection by means of self-assembled DNA-protein conjugates. Nucleic. Acids. Res. 31(16),e90.
- 12. Ooi, D.J.; Dzulkurnain, A.; Othman, R.Y.; Lim, S.H.; Harikrishna, J.A. (2006). Use of superparamagnetic beads for the isolation of a peptide with specificity to cymbidium mosaic virus. J. Virol. Methods. 136(1-2),160-165.
- 13. Park, H.G.; Song, J.Y.; Park, K.H.; Kimb, M.H. (2006). Fluorescence-based assay formats and signal amplication strategies for DNA microarray analysis. Chemical. Engineering. Science. 61(3),954-965.
- 14. Rossi, M.C.; Gori, A.; Zehender, G.; Marchetti, G.; Ferrario, G.; Catozzi, L.; Bandera, A.; Esposti, A.D.; Franzetti, F. (2000). A PCR-colorimetric microwell plate hybridization assay for detection of Mycobacterium tuberculosis and M. avium from culture samples and Ziehl-Neelsen-positive smears. J. Clin. Microbiol 38(5),1772 - 1776.
- 15. Pahwa, R.; Hedau, S.; Jain, S.; Jain, N.; Arora, V.M.; Kumar, N.; Das, B.C. (2005). Assessment of possible tuberculous lymphadenopathy by PCR compared to non-molecular methods. J. Med. Microbiol 54(9),873-878.
- 16. Parsons, L.M.; Brosch, R.; Cole, S.T.; Somoskövi, A.; Loder, A.; Bretzel, G.; Van Soolingen, D.; Hale, Y.M.; Salfinger, M. (2002). Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol 40(7),2339-2345.
- 17. Sener, A.G.; Kurultay, N.; Afsar, I. (2008). Evaluation of the results of Mycobacterium tuberculosis direct test (MTD) and Mycobacterial culture in urine samples. Braz. J. Microbiol. 39(4),673-675.
- 18. Soo, P.C.; Horng, Y.T.; Hsueh, P.R.; Shen, B.J.; Huang, C.C.; Lai, H.C. (2006). Direct and simultaneous identification of Mycobacterium tuberculosis complex (MTBC) and Mycobacterium tuberculosis (MTB) by rapid multiplex nested PCR-ICT assay. J. Microbiol. Methods. 66(3),440-448.
- 19. Steingart, K.R.; Ng, V.; Henry, M.; Hopewell, P.C.; Ramsay, A.; Cunningham, J.; Urbanczik, R.; Perkins, M.D.; Aziz, M.A.; Pai, M. (2006). Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet. Infect. Dis 6(10),664-674.
- 20. Seth, P.; Ahuja, G.K.; Bhanu, N.V.; Behari, M.; Bhowmik, S.; Broor, S.; Dar, L.; Chakraborty, M. (1996). Evaluation of polymerase chain reaction for rapid diagnosis of clinically suspected tuberculous meningitis. Tuber. Lung. Dis. 77(4),353-357.
- 21. Shawar, R.M.; el-Zaatari, F.A.; Nataraj, A.; Clarridge, J.E. (1993). Detection of Mycobacterium tuberculosis in clinical samples by two-step polymerase chain reaction and nonisotopic hybridization methods. J. Clin. Microbiol. 31(1),61-65.
- 22. Verstijnen, C.P.; Ly, H.M.; Polman, K.; Richter, C.; Smits, S.P.; Maselle, S.Y.; Peerbooms, P.; Rienthong, D.; Montreewasuwat, N.; Koanjanart, S. (1991). Enzyme-linked immunosorbent assay using monoclonal antibodies for identification of mycobacteria from early cultures. J. Clin. Microbiol 29(7),1372-1375.
- 23. Wacker, R.; Ceyhan, B.; Alhorn, P.; Schueler, D.; Lang, C.; Niemeyer, C.M. (2007). Magneto immuno-PCR: a novel immunoassay based on biogenic magnetosome nanoparticles. Biochem. Biophys. Res. Commun. 357(2),391-396.
- 24. Wilchek, M.; Bayer, E.A.; Livnah, O. (2006). Essentials of biorecognition: the (strept)avidin-biotin system as a model for protein-protein and protein-ligand interaction. Immunol. Lett 103(1),27-32.
- 25. Yam, W.C.; Cheng,V.C.; Hui, W.T.; Wang, L.N.; Seto, W.H.; Yuen, K.Y. (2004). Direct detection of Mycobacterium tuberculosis in clinical specimens using single-tube biotinylated nested Polymerase chain reaction-enzyme linked immunoassay (PCR-ELISA). Diagn. Microbiol. Infect. Dis 48(4),271-275.
Publication Dates
-
Publication in this collection
21 Dec 2011 -
Date of issue
Sept 2011
History
-
Accepted
14 Mar 2011 -
Reviewed
14 Sept 2010 -
Received
23 Dec 2009