Abstract
Growth and nitrilase production by recombinant Escherichia coli cells harbouring pET 21 (b) plasmid, for the expression of Pseudomonas putida nitrilase were improved using response surface methodology. Central composite design was used for obtaining ideal concentration of critical medium components which include fructose, tryptone, yeast extract and lactose. The optimal values for the concentration of fructose, tryptone, yeast extract and lactose were found to be 1.13, 2.26, 3.25 and 0.9 % (w/v), respectively. Here, fructose served as carbon source for the growth while lactose was preferably used as inducer for the expression of foreign protein. Yeast extract in the medium was used as a growth promoter while tryptone was added as a major nitrogen source. Using this optimized medium, an experimental growth of 6.67 (OD at 600 nm) and nitrilase activity of 27.13 U/ml was achieved. This approach for medium development led to an enhancement of the growth and enzyme activity by 1.4 and 2.2 times, respectively, as compared to the un-optimized medium.
Biocatalysis; Recombinant E. coli; Nitrilase; Response surface methodology; Central composite design
INDUSTRIAL MICROBIOLOGY
Response surface methodology of nitrilase production by recombinant Escherichia coli
Sachin DubeyI,II; Amit SinghI; Uttam C. BanerjeeI,* * Corresponding Author. Mailing address: Biocatalysis Laboratory, Department of Pharmaceutical Technology, National Institute of Pharmaceutical Education and Research, Sector 67, S.A.S. Nagar-160 062, Punjab, India.; Tel.: +91 172 2214682-87 Fax: +91 172 2214692.; E-mail: ucbanerjee@niper.ac.in
IBiocatalysis Laboratory, Department of Pharmaceutical Technology, National Institute of Pharmaceutical Education and Research, Sector 67, S.A.S. Nagar-160 062, Punjab, India
IISchool of Pharmaceutical Sciences, University of Geneva & University of Lausanne, 30 Quai Ernest Ansermet, 1211 Geneva, Switzerland
ABSTRACT
Growth and nitrilase production by recombinant Escherichia coli cells harbouring pET 21 (b) plasmid, for the expression of Pseudomonas putida nitrilase were improved using response surface methodology. Central composite design was used for obtaining ideal concentration of critical medium components which include fructose, tryptone, yeast extract and lactose. The optimal values for the concentration of fructose, tryptone, yeast extract and lactose were found to be 1.13, 2.26, 3.25 and 0.9 % (w/v), respectively. Here, fructose served as carbon source for the growth while lactose was preferably used as inducer for the expression of foreign protein. Yeast extract in the medium was used as a growth promoter while tryptone was added as a major nitrogen source. Using this optimized medium, an experimental growth of 6.67 (OD at 600 nm) and nitrilase activity of 27.13 U/ml was achieved. This approach for medium development led to an enhancement of the growth and enzyme activity by 1.4 and 2.2 times, respectively, as compared to the un-optimized medium.
Key words: Biocatalysis; Recombinant E. coli; Nitrilase; Response surface methodology; Central composite design.
INTRODUCTION
Recent development in molecular biology techniques have helped many fields to flourish and grow faster, this includes biocatalysis as well. Quite often the selected biocatalyst doesn't have desired characteristics and can not be used for industrial purposes. On the other hand the demand of chiral compounds has been increased rapidly over the years (8). This demand can be catered by biocatalysts cloned in user friendly expression system. One of the industrially important building block materials includes nitrile compounds as they can be converted into high value acids and amides (23). Chiral purity in these conversations is often desirable and can be obtained using specific nitrilases. Moreover, nitrilases are expected to gain immense importance as useful biocatalyst for organic chemical processing, because this environment-friendly bioconversion allows clean and mild synthesis with high selectivity and good yield. Recently, we have isolated some new bacterial strains, which convert racemic mandelonitrile to (R)-(-)-mandelic acid (an important chiral building block) with high enantiomeric excess (11). Among them Pseudomonas putida showed comparatively higher yield with excellent ee (99.98%) in shortest possible time. The nitrilase of this organism is also highly stable under operational conditions. Yamamoto et al. first reported (R)-(-)-mandelic acid production with 91% yield and 100% ee (27). Diversa corporation (California, USA) created metagenomic library of nitrilases, 27 of them yielded (R)-(-)-mandelic acid with >90% ee (7). Based on the desirable characteristics of the nitrilase, the gene from P. putida was cloned in E. coli BL 21 using pET 21b plasmid (2).
Optimization of culture conditions resulted in the enhancement of nitrile hydrolyzing activity (16, 17, 24, 25) indicating the role of various culture conditions in the formation of desired enzyme. We used recombinant microorganism under the tight control of T7 promoter, which needed an inducer for expression. Lactose was used as inducer instead of costlier IPTG for the higher expression level of nitrilase. The composition of the fermentation medium plays an important role in the expression of the enzyme catalyzing the desired reaction. It was necessary to study the interaction between lactose and other media components in determining the growth and enzyme production by the recombinant E. coli cells. The study of critical medium components by conventional way i.e. varying one parameter at a time and keeping the other constant; is time consuming and does not allow us to study the effect of interaction of various parameters (6). To overcome this difficulty, response surface methodology (RSM) was employed (1, 20). RSM and factorial design are important tools to study the effect of both the primary factors and their mutual interactions (5, 14). Central composite design (CCD) is a well established, widely used statistical technique for determining the key factors from a large number of medium components by a small number of experiments. Experimental designs have been commonly used for the optimization of multiple variables with minimum number of experiments (10, 13, 15, 19, 26). In this investigation, we tried to improve cell growth as well as nitrilase production of recombinant E. coli by studying various medium components using central composite design.
MATERIALS AND METHODS
Microorganism and culture conditions
The microorganism used in the present work was a recombinant E. coli cell harbouring nitrilase gene in pET 21b(+) (Novagen, Madison, USA) expression vector (2). Pseudomonas putida MTCC 5110 was earlier screened and isolated as a nitrilase producer for the enantioselective hydrolysis of mandelonitrile to (R)-mandelic acid (3). E. coli BL21 (DE3) (Novagen, Madison, USA) was used for the expression of the P. putida nitrilase gene. The stock culture was maintained on Luria Bertani plates containing ampicillin. The microorganism was initially grown at 37 ºC for 16 h in a medium of the following composition: yeast extract (10 g/l), tryptone (16 g/l) and sodium chloride (5 g/l). This inoculum (10%, v/v) was transferred to the production medium and grown for 4 h at 37 ºC in a rotary shaker (200 rpm). Lactose was used as inducer for the expression of nitrilase enzyme.
Estimation of growth
The growth of the organism was estimated by taking 1 ml sample and measuring the optical density (OD) at 600 nm by UV-visible spectrophotometer (Beckman DU 7400, U.S.A.) against water as the blank.
Enzyme activity
The standard reaction mixture (2 ml) consisted of wet cell paste (10 mg) suspended in phosphate buffer (100 mM, pH 7.5). Mandelonitrile (5 mM) was added to initiate the reaction and the mixture was incubated in a water bath maintained at 37 ºC for 20 min. The amount of ammonia produced in the enzymatic reaction was estimated by fluorimetric method (4).
Experimental design and data analysis
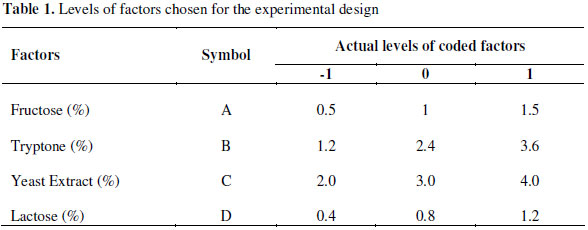
Four experimental factors; fructose, tryptone, yeast extract and lactose were chosen for the enhancement of growth and enzyme production by recombinant E. coli. They were found to have the most significant effect on both growth and enzyme production as determined during the initial studies. Response surface methodology using a four factor, three level central composite design was used to study the response of four variables. A 24 factorial design was generated in order to study the effects of fructose, lactose, yeast extract and tryptone concentration. The growth and enzyme activity after 4 h of induction were studied as responses, as there was no significant increase in the productivity after this time and often 4 h of expression was considered as an ideal time for pET expression system. This criterion was used in all the experiment designs. The statistical analysis of the results was performed by using Design Expert ver. 6.0.9 statistical software (Stat-Ease Inc, Minneapolis, MN). The growth and enzyme activity were analyzed using the analysis of variance (ANOVA) combined with the Fischer test to evaluate if a given term has a significant effect (p< 0.05). The optimum levels of the variables were obtained by graphical and numerical analysis using Design Expert program.
RESULTS AND DISCUSSION
Optimization of culture conditions by central composite design
In order to enhance the growth and enzyme production by a statistically based experimental design, we have chosen fructose, tryptone, yeast extract and lactose as important process parameters. A central composite design (CCD) with three coded levels for all the four factors (fructose (A), tryptone (B), yeast extract (C) and lactose (D)) were used for this purpose. The levels of variables for the CCD were based on the preliminary results. The range of the variables is given in Table 1. The experimental designs and the results obtained for growth and enzyme production are presented in Table 2. The experimental results of the CCD were fitted with a second order polynomial equation. The values of regression coefficients were calculated and the fitted equations (in terms of coded values) for predicting growth (yg) and enzyme activity (yEA) were found to be as:
ŷg = + 6.749 + 0.407*A + 0.137*B + 0.194*C + 0.101*D - 0.426*A2 - 0.413*B2 - 0.392*C2 - 0.300*D2 - 0.069*A*B - 0.075*A*C + 0.057*A*D + 0.035*B*C + 0.397*B*D + 0.065*C*D
ŷEA = + 26.638 - 0.621*A + 0.604*B + 0.482*C + 2.090*D - 1.196*A2 - 1.423*B2 - 1.315*C2 - 3.721*D2 + 0.775*A*B + 0.075*A*C - 0.091*A*D + 0.397*B*C - 2.439*B*D +0.048*C*D
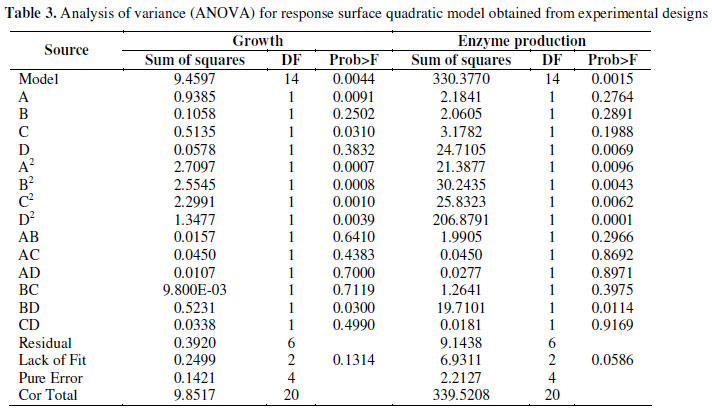
where A, B, C, D represent the concentrations of fructose, tryptone, yeast extract and lactose, respectively (Table 3).
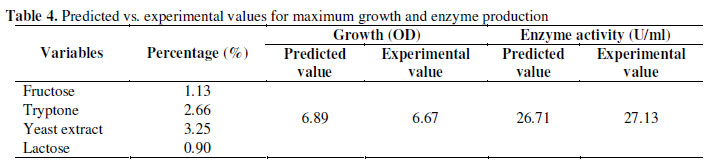
Table 3 shows that the lack of fit and residual values were in range for both growth (Sum of squares: 0.2499 and 0.3920 respectively) and enzyme activity (Sum of squares: 6.9311 and 9.1438 respectively), thus suggesting the validity of the model. The growth and enzyme activity, as predicted by the final quadratic model along with the corresponding observed values, are given in Table 4. Comparison of these values indicated that there was an excellent agreement between the predicted and experimental data.
The location of optimum, obtained by the differentiation of the quadratic model, for achieving maximum growth and enzyme production was A=1.13, B=2.26, C=3.25, D=0.9. The calculated optimal growth and enzyme production corresponding to these values were 6.89 (OD at 600 nm) and 26.71 U/ml. This concentration was the maximum value bounded by the experimental values. To confirm the model accuracy for predicting maximum growth and nitrilase production, additional experiments in triplicates using the optimized medium composition were performed. The triplicate experiments yielded an average maximum growth of 6.67 (OD at 600 nm) and enzyme activity of 27.13 U/ml. The good agreement between the predicted and experimental results verified the validity of the model and the existence of the optimal point.
The response surfaces shown in Fig. 1 were based on the final model, holding two variables constant at their optimum level, while varying the other two within their experimental range. As clear from Fig. 1 (c), the maximum response for enzyme production (27.13 U/ml) occurred when lactose was used in a concentration of 0.9%. On the either side of the optima, the enzyme production decreased considerably. This suggested that the inducer (lactose) concentration in the medium had a very significant effect on the enzyme production. Lactose being natural inducer for Lac operon is not commercially applied because of its instability to resist metabolism. IPTG is used for the expression of recombinant protein under pET expression system but its cost is major hindrance in large scale process development. Thus the use of lactose with the same level of expression was a better option. Here we have shown that 0.9 % lactose along with fructose as an additional carbon source at a concentration of 1.33 % was ideal for maximum nitrilase expression. The concentration of fructose also played an important role. Growth of cells was also found to be maximum at the same concentrations of lactose and fructose (Fig. 1a). The idea of having higher fructose concentration in the medium was to allow the microbes to use fructose for growth and enzyme production while lactose was preferably used as inducer for the expression of foreign proteins. The same was evident from the present experiment where the response varied noticeably at different levels of fructose along the axis suggesting that there was a considerable interaction between these two factors. Similar findings have been reported earlier by many workers. Kotik et. al. (12) have demonstrated the fed-batch culture using lactose and fructose for the production of pyranose oxidase, while Ramchuran et. al. (21) and Gombert et. al. (9) have used glucose as carbon source and lactose as inducer. In another report by Studier (22), the phenomena of auto-induction was explained, while Nair et. al. (18) have shown the self induction of enzyme activity by medium components. The response surfaces shown by Figs. 1 (b) and 1 (d) depicted the combined effect of yeast extract and tryptone in the medium on the growth and enzyme production. Yeast extract was used as a growth promoter while tryptone was used as a major nitrogen source. The growth and enzyme production were maximum when the concentration of yeast extract and tryptone were 3.25 and 2.66 %, respectively.
CONCLUSION
With the growing acceptance for the use of statistical experimental designs in biotechnology, many microbial enzyme productions are improved using statistical approach. This study allowed exploring the culture conditions for the production of nitrilase, approximately optimum of growth and enzyme production was achieve with overall 2.2 fold increase in nitrilase production and 1.4 fold increase in growth. Based on the results, the composition of the fermentation medium for achieving maximum growth and nitrilase activity by recombinant E. coli, consisting of fructose, tryptone, yeast extract and lactose in the concentration of 1.13, 2.26, 3.25 and 0.9%, respectively, was proposed. To the best of our knowledge, there is no report on the application of response surface methodology for the enhancement of nitrilase activity using lactose as inducer.
ACKNOWLEDGEMENT
SD and AS gratefully acknowledges Department of Biotechnology, Government of India for providing research fellowship (M. Tech. Biotechnology Program, NIPER).
REFERENCES
1. Adinarayana, K.; Ellaiah, P. (2002) Response surface optimization of the critical medium components for the production of alkaline protease by a newly isolated Bacillus sp. J. Pharm. Sci. 5,272-278.
2. Banerjee, A.; Dubey, S.; Kaul, P.; Barse, B.; Piotrowski, M.; Banerjee, U. C. (2009) Enantioselective Nitrilase from Pseudomonas putida: Cloning, Heterologous Expression, and Bioreactor Studies. Mol. Biotechnol. 41,35-41.
3. Banerjee, A.; Kaul, P.; Sharma, R.; Banerjee, U.C. (2003) A High-Throughput Amenable Colorimetric Assay for Enantioselective Screening of Nitrilase-Producing Microorganisms Using pH Sensitive Indicators. J. Biomol. Screening. 8,559-565.
4.Banerjee, A.; Sharma, R.; Banerjee, U.C. (2003) A rapid and sensitive fluorometric assay method for the determination of nitrilase activity. Biotech. Appl. Biochem. 37,289-293.
5. Box, G.; Hunter, W.; Hunter, J. (1958) Statistics for experiments. Wiley, New York.
6. Cochran, W.G.; Cox, G.M. (1992) Experimental designs, Wiley, New York.
7. DeSantis, G.; Zhu, Z.; Greenberg, W.A.; Wong, W.A.K.; Chaplin, J.; Hanson, S.R.; Farwell, B.; Nicholson, L.W.; Rand, C.L; Weiner, D.P.; Robertson, D.E.; Burk, M.J. (2002) An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J. Am. Chem. Soc. 124(31),9024-9025.
8. Faber, K. (2000) Biotransformations in Organic Chemistry. Springer-Verlag, Berlin.
9. Ferracini-Santos, L.: Sato H.H. (2009) Production of alkaline protease from Cellulosimicrobium cellulans. Braz. J. Microbiol. 40,54-60.
10. Gombert, A.K.; Kilikian, B.V. (1998) Recombinant gene expression in Escherichia coli cultivation using lactose as inducer. J. Biotechnol. 60,47-54.
11. Kalil, S.J.; Maugeri, F.; Rodrigues, M.I. (2000) Response surface analysis and simulation as a tool for bioprocess design and optimization. Process Biochem. 35,539-550.
12. Kaul, P.; Banerjee, A.; Mayilraj, S.; Banerjee, U. C. (2004) Screening for enantioselective nitrilases: kinetic resolution of racemic mandelonitrile to (R)-(-)-mandelic acid by new bacterial isolates. Tetrahedron Asymmetr. 15,207-211.
13. Kawaguti, H.K.; Manrich, E.; Fleuri, L.F.; Sato, H.H. (2005) Production of glucosyltransferase by Erwinia sp. Using experimental design and response surface methodology. Braz. J. Microbiol. 36,227-234.
14. Kotik, M.; Kocanova, M.; Maresova, H.; Kyslik, P. (2004) High-level expression of a fungal pyranose oxidase in high cell-density fed-batch cultivations of Escherichia coli using lactose as inducer. Protein. Expres. Purif. 36,61-69.
15. Melo, I.R.; Pimental, M.F.; Lopes, C.E.; Calazans, G.M.T. (2007) Application of fractional factorial design to levan production by Zymomonas mobilis. Braz. J. Microbiol. 38,45-51.
16. Montgomery, D.C. (2001) Design and analysis of experiments. Wiley, New York.
17. Nagasawa, T.; Kobayashi, M.; Yamada, H. (1988) Optimum culture conditions for the production of benzonitrilase by Rhodococcus rhodochrous J1. Arch. Microbiol. 150,89-94.
18. Nagasawa, T.; Nakamura, T.; Yamada, H. (1990)  -Caprolactam, a new powerful inducer for the formation of Rhodococcus rhodochrous J1 nitrilase. Arch. Microbiol. 155,13-17.
-Caprolactam, a new powerful inducer for the formation of Rhodococcus rhodochrous J1 nitrilase. Arch. Microbiol. 155,13-17.
19. Nair, R.; Salvi, P.; Banerjee, S.; Raiker, V.A.; Bandyopadhyay, S.; Soorapaneni, S.; Kotwal, P.; Padmanabhan, S. (2009) Yeast extract mediated autoinduction of lacUV5 promoter: an insight. New Biotechnol. 26(6),282-288.
20. Pacheco, G.J.; Ciapina, E.M.P.; Gomes, E.B.; Pereira Junior, N. (2010) Biosurfactant production by Rhodococcus erythropolis and its application to oil removal. Braz. J. Microbiol. 41:685-693.
21. Pansuriya, R.C.; Singhal R.S. (2010) Response surface methodology for optimization of production of lovastatin by solid state fermentation. Braz. J. Microbiol. 41,164-172.
22. Ramchuran, S.O.; Holst, O.; Karlsson, E.N. (2005) Effect of postinduction nutrient feed composition and use of lactose as inducer during production of thermostable xylanase in Escherichia coli glucose-limited fed-batch cultivations. J. Biosci. Bioeng. 99(5),477-484.
23. Studier, F.W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein. Expres. Purif. 41,207-234.
24. Sugai, T.; Yamazaki, T.; Yokoyama. M.; Ohta, H. (1997) Biocatalysis in
25. organic synthesis: the use of nitrile and amide hydrolysing microorganisms. Biosci. Biotechnol. Biochem. 61,1419-1427.
26. Watanabe, I.; Satoh, Y.; Enomoto, K.; Seki, S.; Sakshita, K. (1987) Optimal conditions for cultivation of Rhodococcus sp. N774 and conversion of acrylamide by resting cells. Agric. Biol. Chem. 51,3201-3206.
27. Yamada, H.; Ryuno, K.; Nagasawa, T.; Enomoto, K.; Watanabe, I. (1986) Optimum culture conditions for production of nitrile hydratase by Pseudomonas chlororaphis B23. Agric. Biol. Chem. 50,2859-2865.
28. Yamamoto, K.; Oishi, K.; Fujimatsu, I.; Komatsu, K. (1991) Production of R-(-)-mandelic acid from mandelonitrile by Alcaligenes faecalis ATCC 8750. Appl. Environ. Microbiol. 57(10),3028-3032.
Submitted: July 18, 2009; Returned to authors for corrections: November 05, 2010; Approved: March 14, 2011.
- 1. Adinarayana, K.; Ellaiah, P. (2002) Response surface optimization of the critical medium components for the production of alkaline protease by a newly isolated Bacillus sp. J. Pharm. Sci. 5,272-278.
- 2. Banerjee, A.; Dubey, S.; Kaul, P.; Barse, B.; Piotrowski, M.; Banerjee, U. C. (2009) Enantioselective Nitrilase from Pseudomonas putida: Cloning, Heterologous Expression, and Bioreactor Studies. Mol. Biotechnol 41,35-41.
- 3. Banerjee, A.; Kaul, P.; Sharma, R.; Banerjee, U.C. (2003) A High-Throughput Amenable Colorimetric Assay for Enantioselective Screening of Nitrilase-Producing Microorganisms Using pH Sensitive Indicators. J. Biomol. Screening. 8,559-565.
- 4.Banerjee, A.; Sharma, R.; Banerjee, U.C. (2003) A rapid and sensitive fluorometric assay method for the determination of nitrilase activity. Biotech. Appl. Biochem. 37,289-293.
- 5. Box, G.; Hunter, W.; Hunter, J. (1958) Statistics for experiments. Wiley, New York.
- 6. Cochran, W.G.; Cox, G.M. (1992) Experimental designs, Wiley, New York.
- 7. DeSantis, G.; Zhu, Z.; Greenberg, W.A.; Wong, W.A.K.; Chaplin, J.; Hanson, S.R.; Farwell, B.; Nicholson, L.W.; Rand, C.L; Weiner, D.P.; Robertson, D.E.; Burk, M.J. (2002) An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J. Am. Chem. Soc. 124(31),9024-9025.
- 8. Faber, K. (2000) Biotransformations in Organic Chemistry. Springer-Verlag, Berlin.
- 9. Ferracini-Santos, L.: Sato H.H. (2009) Production of alkaline protease from Cellulosimicrobium cellulans. Braz. J. Microbiol 40,54-60.
- 10. Gombert, A.K.; Kilikian, B.V. (1998) Recombinant gene expression in Escherichia coli cultivation using lactose as inducer. J. Biotechnol 60,47-54.
- 11. Kalil, S.J.; Maugeri, F.; Rodrigues, M.I. (2000) Response surface analysis and simulation as a tool for bioprocess design and optimization. Process Biochem. 35,539-550.
- 12. Kaul, P.; Banerjee, A.; Mayilraj, S.; Banerjee, U. C. (2004) Screening for enantioselective nitrilases: kinetic resolution of racemic mandelonitrile to (R)-(-)-mandelic acid by new bacterial isolates. Tetrahedron Asymmetr 15,207-211.
- 13. Kawaguti, H.K.; Manrich, E.; Fleuri, L.F.; Sato, H.H. (2005) Production of glucosyltransferase by Erwinia sp. Using experimental design and response surface methodology. Braz. J. Microbiol 36,227-234.
- 14. Kotik, M.; Kocanova, M.; Maresova, H.; Kyslik, P. (2004) High-level expression of a fungal pyranose oxidase in high cell-density fed-batch cultivations of Escherichia coli using lactose as inducer. Protein. Expres. Purif 36,61-69.
- 15. Melo, I.R.; Pimental, M.F.; Lopes, C.E.; Calazans, G.M.T. (2007) Application of fractional factorial design to levan production by Zymomonas mobilis. Braz. J. Microbiol 38,45-51.
- 16. Montgomery, D.C. (2001) Design and analysis of experiments. Wiley, New York.
- 17. Nagasawa, T.; Kobayashi, M.; Yamada, H. (1988) Optimum culture conditions for the production of benzonitrilase by Rhodococcus rhodochrous J1. Arch. Microbiol 150,89-94.
- 18. Nagasawa, T.; Nakamura, T.; Yamada, H. (1990) -Caprolactam, a new powerful inducer for the formation of Rhodococcus rhodochrous J1 nitrilase. Arch. Microbiol. 155,13-17.
- 19. Nair, R.; Salvi, P.; Banerjee, S.; Raiker, V.A.; Bandyopadhyay, S.; Soorapaneni, S.; Kotwal, P.; Padmanabhan, S. (2009) Yeast extract mediated autoinduction of lacUV5 promoter: an insight. New Biotechnol 26(6),282-288.
- 20. Pacheco, G.J.; Ciapina, E.M.P.; Gomes, E.B.; Pereira Junior, N. (2010) Biosurfactant production by Rhodococcus erythropolis and its application to oil removal. Braz. J. Microbiol 41:685-693.
- 21. Pansuriya, R.C.; Singhal R.S. (2010) Response surface methodology for optimization of production of lovastatin by solid state fermentation. Braz. J. Microbiol 41,164-172.
- 22. Ramchuran, S.O.; Holst, O.; Karlsson, E.N. (2005) Effect of postinduction nutrient feed composition and use of lactose as inducer during production of thermostable xylanase in Escherichia coli glucose-limited fed-batch cultivations. J. Biosci. Bioeng. 99(5),477-484.
- 23. Studier, F.W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein. Expres. Purif 41,207-234.
- 26. Watanabe, I.; Satoh, Y.; Enomoto, K.; Seki, S.; Sakshita, K. (1987) Optimal conditions for cultivation of Rhodococcus sp. N774 and conversion of acrylamide by resting cells. Agric. Biol. Chem. 51,3201-3206.
- 27. Yamada, H.; Ryuno, K.; Nagasawa, T.; Enomoto, K.; Watanabe, I. (1986) Optimum culture conditions for production of nitrile hydratase by Pseudomonas chlororaphis B23. Agric. Biol. Chem. 50,2859-2865.
- 28. Yamamoto, K.; Oishi, K.; Fujimatsu, I.; Komatsu, K. (1991) Production of R-(-)-mandelic acid from mandelonitrile by Alcaligenes faecalis ATCC 8750. Appl. Environ. Microbiol. 57(10),3028-3032.
Publication Dates
-
Publication in this collection
21 Dec 2011 -
Date of issue
Sept 2011
History
-
Received
18 July 2009 -
Reviewed
05 Nov 2010 -
Accepted
14 Mar 2011