Abstract
Bacteria associated with the parthenogenetic troglobiont sand fly Deanemyia maruaga were characterized by sequencing cloned 16S rDNA PCR products. Eleven novel partial 16S rDNA sequences, with varying degrees of similarity to Actinobacteria, were identified. None of the sequences identified had homology to those known from parthenogenesis-inducing bacteria.
Actinobacteria; bat guano; parthenogensis; sand flies; troglobiont
16S rRNA gene-based identification of microbiota associated with the parthenogenetic troglobiont sand fly Deanemyia maruaga (Diptera, Psychodidae) from central Amazon, Brazil
Katianne Barbosa Alves de SousaI; Túllio Romão Ribeiro da SilvaI; Ronildo Baiatone AlencarII; Luke Anthony BatonIII; Felipe Gomes NavecaI; Paloma Helena Fernandes ShimabukuroI,III
IInstituto Leônidas e Maria Deane, Fiocruz/Amazônia, Manaus, AM, Brazil
IICoordenação de Pesquisas em Entomologia, Instituto Nacional de Pesquisas da Amazônia, Manaus, AM, Brazil
IIICentro de Pesquisas René Rachou, Fiocruz/Minas, Belo Horizonte, MG, Brazil
Send correspondence Send correspondence to: P.H.F. Shimabukuro Centro de Pesquisas René Rachou, Fiocruz/Minas Avenida Augusto de Lima 1715, Barro Preto 31190-002 Belo Horizonte, MG, Brazil E-mail: phfs@yahoo.com
ABSTRACT
Bacteria associated with the parthenogenetic troglobiont sand fly Deanemyia maruaga were characterized by sequencing cloned 16S rDNA PCR products. Eleven novel partial 16S rDNA sequences, with varying degrees of similarity to Actinobacteria, were identified. None of the sequences identified had homology to those known from parthenogenesis-inducing bacteria.
Key words: Actinobacteria, bat guano, parthenogensis, sand flies, troglobiont.
Sand flies are dioecious, holometabolous insects responsible for transmission of Leishmania species, bacteria and viruses to humans and non-human animals (Rangel and Lainson, 2011). The ground-dwelling larvae feed on decaying organic matter present in the forest soil, animal burrows and rock crevices (Rangel and Lainson, 2011). In contrast to most known sand fly species, the non-vector Deanemyia maruaga is a parthenogenetic troglobiont (Alves et al., 2008). It has been described from only a single cave in the central Amazon, Brazil, within which it completes its entire life-cycle (Alves et al., 2008). Under laboratory conditions, D. maruaga has been shown to be autogenous, i.e. it does not require a bloodmeal for egg development, which implies that feeding, sufficient for reproduction, only occurs during the larval stages. Only female adults have so far been captured, suggesting that this species is parthenogenetic (Alves et al., 2008, 2011). Parthenogenesis has been described in most insect orders and occurs by a variety of mechanisms (Normark and Kirkendall, 2009), but has only previously been reported for one other sand fly species, Pintomyia mamedei (Brazil and Oliveira, 1999). Recently, the occurrence of parthenogenesis in insects has been associated with the presence of various maternally-inherited bacteria (Hagimori et al., 2006; Werren et al., 2008; Zchori-Fein and Perlman, 2004). Here, we describe the bacteria associated with D. maruaga using the culture-independent method of sequencing cloned 16S rDNA PCR products.
The larvae and adults of D. maruaga were collected from the Refúgio do Maruaga cave, located in the municipality of Presidente Figueiredo, Amazonas state, Brazil (02º03'02" S, 59º57'48" W). Two collections were carried out in 2008/2009: one in the rainy season (March) and another in the dry season (November). In total, approximately 40 kg of guano were collected and immature sand flies were separated in the laboratory using the flotation method (Hanson, 1961). Adults of D. maruaga were collected using 10 CDC light traps distributed along the cave 5 m apart from each other, and approximately 2 m high. The insects were surface sterilized in 70% ethanol and then rinsed in sterile phosphate buffered saline (PBS) (Lindh et al., 2008; Volf et al., 2002). To detect and identify the bacteria that may be present in the sand fly larvae and adults, DNA was isolated, and a 586 bp partial fragment was amplified by touchdown PCR, cloned and sequenced as described elsewhere (Muyzer et al., 1993; Muyzer et al., 1998). Nucleotide sequences corresponding to 16S rRNA gene fragments have been deposited in GenBank under accession numbers JN040717 to JN040727.
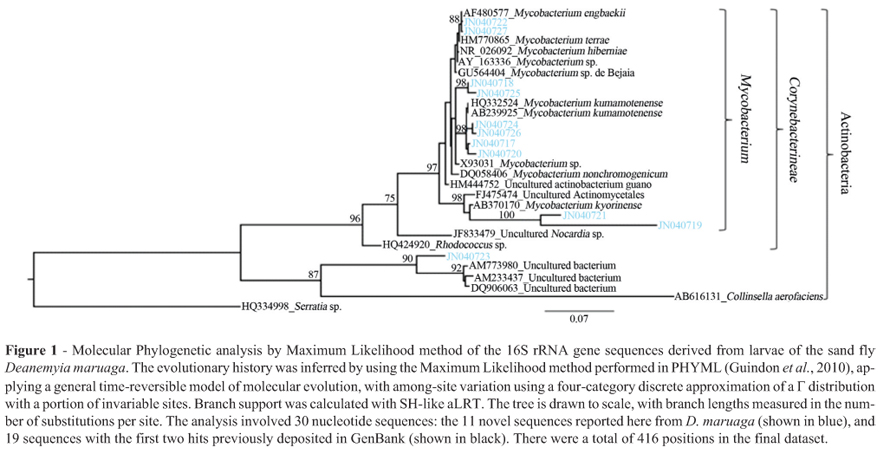
Phylogenetic analyses were carried out with maximum likelihood (ML) method performed in PHYML (Guindon et al., 2010). The sequences were checked for chimeras using Greengenes NAST server alignment tool (DeSantis et al., 2006) and Bellerophon 3 (Huber et al., 2004). None of the sequences were chimeric.
A total of 11 clones were successfully sequenced, yielding 11 unique partial 16S rRNA gene sequences, isolated from four pooled samples of the larvae of D. maruaga (Table 1 and Figure 1). No 16S rDNA PCR products were obtained from the adults of D. maruaga, even though the positive controls amplified the expected product.
Phylogenetic analysis of the 16S rDNA sequences recovered from the larvae of D. maruaga (Figure 1) showed that ten of our sequences grouped in the genus Mycobacterium, mainly from the nonpathogenic M. terrae complex (Torkko et al., 1998). The remaining sequence (JN040723), also had similarity, albeit relatively low, to previously reported sequences from diverse environmental sources: soil samples (Yarwood et al., 2009), water supplies (Le Dantec et al., 2002), and rhizospheres (Burke et al., 2008; Mirete et al., 2007), respectively. It forms an independent and taxonomically unassigned lineage affiliated with Actinobacteria, also containing several other sequences previously deposited in GenBank obtained from uncultured environmental rhizosphere and soil samples.
The bacteria we found in the larvae of D. maruaga are very distinct from the microbiota previously described from other sand fly species (Dillon et al., 1996; Volf et al., 2002; Gouveia et al., 2008; Hillesland et al., 2008). Most previous studies investigated adults of vector species, which feed on plant juices and vertebrate blood (Gouveia et al., 2008). The bacteria found in these sand flies were pre-dominantly Gram-negative members of the γ-Proteobacteria, and a few Gram-positive Firmicutes (Dillon et al., 1996; Gouveia et al., 2008; Hillesland et al., 2008). In contrast, only Actinobacteria were found in the larvae of D. maruaga, and this class of bacteria, including mycobacteria, has not previously been associated with any species of sand fly.
Larvae of D. maruaga live within, and feed upon, bat guano deposited on the cave floor, and are likely to acquire their microbiota from the bat guano ingested during feeding. Seven of the sequences we report here had BLAST matches to two 16S rDNA sequences previously isolated from bat guano collected in a cave in Slovakia (Hill et al., 2011), although another microbiological survey of bat guano did not report the occurrence of Actinobacteria (Konieczna et al., 2007). Whether the Actinobacteria in the bat guano are derived from the bats (i.e. their defecated intestinal microbiome) or the cave environment (i.e. soil or rock walls) is unclear. Culture-dependent microbiological analysis of the gut content of bats has shown the presence of Actinobacteria from the genus Corynebacterium (Klite, 1965; Heard et al., 1997). However, the presence of Actinobacteria has also been reported from rock wall surfaces and soil samples from other caves through culture-independent methods (Schabereiter-Gurtner et al., 2002, 2004; Zhou et al., 2007), indicating a possible environmental source for the Actinobacteria in bat guano. Bat guano is acidic, and would accordingly provide an ideal, nutrient-rich substrate for acidophilic Actinobacteria (Goodfellow and Williams, 1983).
The absence of amplification of bacterial DNA from the adults of D. maruaga is unexpected, but might be explained by the apparent absence of transtadial passage of bacteria during the larval to adult metamorphosis (Killick-Kendrick, 1979; Lindh et al., 2008; Moll et al., 2001) and the autogeny of D. maruaga (Alves et al., 2011), eliminating the microbial exposures experienced by adults of other sand fly species associated with ingestion of plant juices and bloodfeeding on vertebrate hosts. Another possibility is that the density of bacteria associated with adults of D. maruaga is extremely low, several orders of magnitude lower than that of larvae, and therefore below the detection threshold of our PCR reaction conditions.
The universal eubacterial primers we used in our study should allow the detection of 16S rDNA fragments from the endosymbiotic bacteria associated with the occurrence of parthenogenesis in other insects (e.g. Wolbachia, Rickettsia and Cardinium) (Hagimori et al., 2006; Werren et al., 2008; Zchori-Fein and Perlman, 2004). Since we did not detect sequences from any of these or related bacteria in either the larvae or adults of D. maruaga, another mechanism is probably responsible for the parthenogenesis of this sand fly. The apparent absence of Actinobacteria from the adults of D. maruaga would seem to preclude the possibility that these bacteria cause parthenogenesis.
We would like to thank: Programa Integrado de Pesquisa Científica e Tecnológica/Fundação de Amparo à Pesquisa do Estado do Amazonas for grant no. 2216/08 to PHFS; the Department of Environment and Tourism in Presidente Figueiredo for permission to undertake field work at the Refúgio do Maruaga cave. We are also grateful to Ricardo de Moura Mota and Sílvio Romero Junior for field work assistance.
Submitted: November 24, 2011
Approved: July 2, 2012
All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License CC BY-NC.
- Alves VR, Freitas RA, Barrett T (2008) Lutzomyia maruaga (Diptera, Psychodidae), a new bat-cave sand fly from Amazonas, Brazil. Mem Inst Oswaldo Cruz 103:251-253.
- Alves VR, Freitas RA, Santos FL, Barrett TV (2011) Diversity of sandflies (Psychodidae, Phlebotominae) captured in sandstone caves from Central Amazonia, Brazil. Mem Inst Oswaldo Cruz 106:353-359.
- Brazil RP, Oliveira SM (1999) Parthenogenesis in the sandfly Lutzomyia mamedei (Diptera, Psychodidae). Med Vet Entomol 13:463-464.
- Burke DJ, Dunham SM, Kretzer AM (2008) Molecular analysis of bacterial communities associated with the roots of Douglas fir (Pseudotsuga menziesii) colonized by different ectomycorrhizal fungi. FEMS Microbiol Ecol 65:299-309.
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069-5072.
- Dillon RJ, el Kordy E, Shehata M, Lane RP (1996) The prevalence of a microbiota in the digestive tract of Phlebotomus papatasi Ann Trop Med Parasitol 90:669-673.
- Goodfellow M, Williams ST (1983) Ecology of Actinomycetes. Annu Rev Microbiol 37:189-216.
- Gouveia C, Asensi MD, Zahner V, Rangel EF, Oliveira SM (2008) Study on the bacterial midgut microbiota associated to different Brazilian populations of Lutzomyia longipalpis (Lutz & Neiva) (Diptera, Psychodidae). Neotrop Entomol 37:597-601.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59:307-321.
- Hagimori T, Abe Y, Date S, Miura K (2006) The first finding of a Rickettsia bacterium associated with parthenogenesis induction among insects. Curr Microbiol 52:97-101.
- Hanson WJ (1961) The breeding places of Phlebotomus in Panama (Diptera, Psychodidae). Ann Entomol Soc Am 54:317-322.
- Heard DJ, De Young JL, Goodyear B, Ellis GA (1997) Comparative rectal bacterial flora of four species of flying fox (Pteropus sp.). J Zoo Wildl Med 28:471-475.
- Hill P, Kristufek V, Dijkhuizen L, Boddy C, Kroetsch D, van Elsas JD (2011) Land use intensity controls actinobacterial community structure. Microb Ecol 61:286-302.
- Hillesland H, Read A, Subhadra B, Hurwitz I, McKelvey R, Ghosh K, Das P, Durvasula R (2008) Identification of aerobic gut bacteria from the Kala Azar vector, Phlebotomus argentipes: A platform for potential paratransgenic manipulation of sand flies. Am J Trop Med Hyg 79:881-886.
- Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319.
- Killick-Kendrick R (1979) Biology of Leishmania in phlebotomine sandflies. In: Lumsden WHR, Evans DA (eds) Biology of Kinetoplastida II. Academic Press, London, pp 395-460.
- Klite PD (1965) Intestinal bacterial flora and transit time of three neotropical bat species. J Bacteriol 90:375-379.
- Konieczna I, Durlik M, Winkowski M, Domañski J, Markowski J, Kaca W (2007) Properties of bacterial microflora isolated from bat guano. Med Wet 63:1626-1629.
- Le Dantec C, Duguet JP, Montiel A, Dumoutier N, Dubrou S, Vincent V (2002) Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Appl Environ Microbiol 68:1025-1032.
- Lindh JM, Borg-Karlson AK, Faye I (2008) Transstadial and horizontal transfer of bacteria within a colony of Anopheles gambiae (Diptera, Culicidae) and oviposition response to bacteria-containing water. Acta Trop 107:242-250.
- Mirete S, de Figueras CG, Gonzalez-Pastor JE (2007) Novel nickel resistance genes from the rhizosphere metagenome of plants adapted to acid mine drainage. Appl Environ Microbiol 73:6001-6011.
- Moll RM, Romoser WS, Modrzakowski MC, Moncayo AC, Lerdthusnee K (2001) Meconial peritrophic membranes and the fate of midgut bacteria during mosquito (Diptera, Culicidae) metamorphosis. J Med Entomol 38:29-32.
- Muyzer G, Brinkhoff T, Ntibel U, Santegoeds C, Schifer H, Wawer C (1998) Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans ADL, van Elsas JD, de Bruijn EJ (eds) Molecular Microbial Ecology Manual. Kluwer Academic Publishers, Dordrecht, pp 1-27.
- Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16S ribosomal-RNA. Appl Environ Microbiol 59:695-700.
- Normark BB, Kirkendall RL (2009) Parthenogenesis in Insects and Mites. In: Resh VH, Cardé RT (eds) Encyclopedia of Insects. Academic Press, London, pp 688-756.
- Rangel EF, Lainson R (2011) Ecologia das Leishmanioses. In: Rangel EF, Lainson R (eds) Flebotomíneos do Brasil. Editora Fiocruz, Rio de Janeiro, pp 291-309.
- Schabereiter-Gurtner C, Saiz-Jimenez C, Pinar G, Lubitz W, Rolleke S (2002) Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its Palaeolithic paintings. Environ Microbiol 4:392-400.
- Schabereiter-Gurtner C, Saiz-Jimenez C, Pinar G, Lubitz W, Rolleke S (2004) Phylogenetic diversity of bacteria associated with Paleolithic paintings and surrounding rock walls in two Spanish caves (Llonin and La Garma). FEMS Microbiol Ecol 47:235-247.
- Torkko P, Suutari M, Suomalainen S, Paulin L, Larsson L, Katila ML (1998) Separation among species of Mycobacterium terrae complex by lipid analyses: Comparison with biochemical tests and 16S rRNA sequencing. J Clin Microbiol 36:499-505.
- Volf P, Kiewegova A, Nemec A (2002) Bacterial colonisation in the gut of Phlebotomus duboscqi (Diptera, Psychodidae): Transtadial passage and the role of female diet. Folia Parasitol 49:73-77.
- Werren JH, Baldo L, Clark ME (2008) Wolbachia: Master manipulators of invertebrate biology. Nat Rev Microbiol 6:741-751.
- Yarwood SA, Myrold DD, Hogberg MN (2009) Termination of belowground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. FEMS Microbiol Ecol 70:151-162.
- Zchori-Fein E, Perlman SJ (2004) Distribution of the bacterial symbiont Cardinium in arthropods. Mol Ecol 13:2009-2016.
- Zhou J, Gu Y, Zou C, Mo M (2007) Phylogenetic diversity of bacteria in an earth-cave in Guizhou province, southwest of China. J Microbiol 45:105-112.
Publication Dates
-
Publication in this collection
05 Apr 2013 -
Date of issue
2013
History
-
Received
24 Nov 2011 -
Accepted
02 July 2012