Abstract
A strain of lactic acid bacteria, Leuconostoc lactis, was isolated from the intestinal tract of black porgy, Sparus macrocephalus, and identified by conventional biochemical characteristics and 16S rDNA gene sequence analysis. The isolated strain had the ability of bile tolerance and resistance to low pH, and survived well in the trypsinase and pepsin solution. But the highly concentrated dose of trypsinase and pepsin affect the viability of the isolated strain. The isolate was resistant to several antibiotics, including Cephalothin, Ceftriaxone, Imipenem and Tobramycin. The isolate could autoaggregate itself and coaggregate with other bacteria in vitro. The autoaggregation percentage increased to 23.29% after 20 h of incubation. The percentage of coaggregation were respectively 31.21%, 29.44%, 10.74%, 16.49%, 24.36%, 24.41% and 20.99% for Vibrio parahaemolyticus, Listeria monocytogenes, Escherichia coli O157, Salmonella typhimurium, Shigella, Staphylococcus aureus and Proteusbacillus vulgaris after 20 h incubation of a mixed suspension. The supernatant of the strain inhibited the growth of several pathogens, such as V.parahaemolyticus, Vibrio harveyi, Vibrio alginolyticus, Staphylococcus aureus, Escherichia coli O157, Salmonella typhimurium, Bacillus subtilis, Proteusbacillus vulgaris and Shigella. These results indicated that the isolate, Leuconostoc lactis, might be an attractive candidate for perspectival strain for probiotics in marine aquaculture.
black porgy; Leuconostoc lactis; lactic acid bacteria; biological characteristic; probiotic
Biological characteristics and probiotic effect of Leuconostoc lactis strain isolated from the intestine of black porgy fish
Wei Zhang; Mingqi Liu; Xianjun Dai
College of Life Sciences, China Jiliang University, Hangzhou, China
Send correspondence to Send correspondence to X. Dai College of Life Sciences, China Jiliang University 258 Xueyuan Street, Xiasha 310018 Hangzhou, People's Republic China E-mail: xjdai@cjlu.edu.cn
ABSTRACT
A strain of lactic acid bacteria, Leuconostoc lactis, was isolated from the intestinal tract of black porgy, Sparus macrocephalus, and identified by conventional biochemical characteristics and 16S rDNA gene sequence analysis. The isolated strain had the ability of bile tolerance and resistance to low pH, and survived well in the trypsinase and pepsin solution. But the highly concentrated dose of trypsinase and pepsin affect the viability of the isolated strain. The isolate was resistant to several antibiotics, including Cephalothin, Ceftriaxone, Imipenem and Tobramycin. The isolate could autoaggregate itself and coaggregate with other bacteria in vitro. The autoaggregation percentage increased to 23.29% after 20 h of incubation. The percentage of coaggregation were respectively 31.21%, 29.44%, 10.74%, 16.49%, 24.36%, 24.41% and 20.99% for Vibrio parahaemolyticus, Listeria monocytogenes, Escherichia coli O157, Salmonella typhimurium, Shigella, Staphylococcus aureus and Proteusbacillus vulgaris after 20 h incubation of a mixed suspension. The supernatant of the strain inhibited the growth of several pathogens, such as V.parahaemolyticus, Vibrio harveyi, Vibrio alginolyticus, Staphylococcus aureus, Escherichia coli O157, Salmonella typhimurium, Bacillus subtilis, Proteusbacillus vulgaris and Shigella. These results indicated that the isolate, Leuconostoc lactis, might be an attractive candidate for perspectival strain for probiotics in marine aquaculture.
Key words: black porgy, Leuconostoc lactis, lactic acid bacteria, biological characteristic, probiotic.
Introduction
With the development of commercial scale aquaculture, it has become apparent that disease can be a significant limiting factor for the fish in sea water. Chemotherapeutic agents, i.e. antibiotics, could lead to the evolution of resistance among pathogenic bacteria (Angulo 2000; Balcázar et al., 2006) and should be minimized in the prevention of disease of marine fish. Probiotic, a kind of live microorganism, may reduce the incidence of disease or lessen the severity of disease outbreaks as the substitution for chemotherapeutic agents (Reid et al., 2003).
Probiotics are defined as microbial dietary adjuvants that beneficially affect the host by modulating mucosal and systemic immunity, as well as improving nutritional and microbial balance in the intestinal tract. Lactic acid bacteria (LAB) is generally used as the probiotics in aquaculture, and have not been found to cause infectious disease for fishes (Verschuere et al., 2000). The mechanisms of LAB used as probiotic include the production of inhibitory substances against pathogens, competition for essential nutrients, adhesion sites and tolerance for pH and bile in stomach-intestinal tract. As a ‘Generally Recognized As Safe' (GRAS) classification, the role of LAB has been extensively studied in the digestive tract of larvae and juvenile fish (Ringø et al., 1998) .
The black porgy, Sparus macrocephalus, had been widely cultured in eastern China for its fast growth rate, readily adaptation to different environment, and better resistance to disease. In the study, a strain of lactic acid bacteria from the intestine of the fishes, had been isolated and identified, and its biological characteristics and probiotic effect were evaluated for the potentially applications in marine aquaculture.
Materials and Methods
Fish and bacteria sampling
Ten healthy black porgies (about 50 g) were collected from a commercial fish farm without antibiotic in the fish feeds at Zhoushan islands, Zhejiang province, China, in June 2010. Each fish was dissected aseptically after capture, and its intestine was taken out and washed in use of sterile 50% artificial seawater (Rei-Sea, Tokyo, Japan). The intestine samples were grinded and serially diluted with sterile artificial seawater (Rei-Sea, Tokyo, Japan). Then solutions were plated onto MRS-agar medium containing 2% agar for selection of lactic acid bacteria. The MRS broth (BD, Franklin Lakes, NJ, USA) was prepared with artificial seawater. The plates were incubated at 30 ºC for 3 days under aerobic conditions. Subsequently, the loopful clones from the MRS-agar medium were inoculated in MRS broth and incubated at 30 ºC in an anaerobic jar.
All isolates were examined for phenotypic properties including Gram reaction, cellular morphology and colony pigmentation. To be tested for gas production, the isolates were inoculated into phenol red broth (Ishida et al., 2000) containing 1% glucose, 1% peptone, 0.5% sodium chloride and 0.0018% phenol red with a Durham tube (pH 6.28), and incubated at 30 ºC for 48 h.
PCR amplification of 16S rDNA sequence and phylogenetic tree
The genomic DNA of the isolated strain was extracted by TaKaRa Mini BEST Bacterial Genomic DNA Extraction Kit Ver.2.0 (Takara, Dalian, China). PCR-mediated amplification primers were respectively a forward primer (5'-GAGCGGATAACAATTTCACACAGG-3') and a reverse primer (5'-CGCCAGGGTTTTCCCAGTC ACGAC-3') and DNA polymerase was Takara Ex Taq (Takara, Dalian, China). The PCR amplifications reaction was carried out in sterile seal tubes with cap locks and performed in the following conditions: initial denaturation of template DNA at 94 ºC for 4 min; then 30 cycles consisting of denaturation at 94 ºC for 30 s, annealing at 55 ºC for 30 s, extension at 72 ºC for 50 s, and a final extension at 72 ºC for 10 min. The PCR-products were purified and ligated into a pMD18-T vector. The inserted DNA was analyzed by an automatic sequencer. The BLAST programs were applied in order to search the sequences similarity in 16S rDNA databases. Sequences were then aligned with GenBank (Stoesser et al., 2001) and phylogenetic trees were drawn after distances had been determined by neighbor-joining algorithm (Saitou et al., 1987) using the same software.
Effect of pH on survival rate of the isolate
The bacterial suspension of the isolate was serially diluted in sterile PBS and flooded with 100 µL on the surface of MRS agar with the pH of 1.5, 2.5, 3.5 and 4.5, respectively. The MRS agar with pH 5.5 was used as control. The plates were incubated for 24 h at 30 ºC in an anaerobic condition. After incubation, the viable bacteria on every plate were counted. For each assay in this study, triplicate was performed to correct for intra-assay variation.
Tolerance for the bile salt
The bacterial were prepared with sterile PBS containing 0, 1, 2, 3, 4 (g/L) bile, and then incubated for 6 h at 30 ºC in an anaerobic condition. After incubation, the suspensions were serially diluted in sterile PBS and counts of viable bacteria were determined by plate count using MRS agar (De Man et al., 1960) incubated in an anaerobic condition at 30 ºC.
Survival in trypsinase and pepsin juice in vitro
The isolate was evaluated by checking the tolerance of the simulated gastric juice and the simulated intestinal juice in vitro. Trypsinase and pepsin were purchased from Shanghai songon Company (songon, Shanghai, China). The bacterial suspension (107 cfu/mL of the isolate) was prepared with sterile PBS containing trypsinase of 1, 3, 5, 7 g/L at pH 6.8, respectively. Tolerance for pepsin juice was determined using a bacterial suspension (107 cfu/mL of the isolate) in sterile PBS containing pepsin of 0, 4, 6, 8,10 g/L respectively at pH 3.0, respectively. After incubation for 6 h at 30 ºC in an anaerobic jar condition, each bacterial suspension of 100 µL was flooded on the surface of the MRS agar and then was counted.
Antibiotic susceptibility of the isolate
The microorganism, Staphylococcus aureus (ATCC29213) strain was employed as reference strain. Antimicrobial susceptibility was studied by employing the method described in reference (Bauer et al., 1966). The minimum inhibitory concentration (MIC) was defined as the lowest concentration of an agent that yielded no growth or a marked change in the appearance of growth, compared to the growth on a control plate. The isolate was cultured in MRS at 30 ºC in an anaerobic jar for 24 hours. The concentration of suspensions was adjusted to 108 cfu/mL and the isolate was disseminated on the surface of MRS agar plates. Antibiotic discs were placed on the surface of the MRS (three discs in each plate) and the plates were incubated for 24 hours at 30 ºC in an anaerobic jar. After the incubation, the diameter of the halos was measured.
Autoaggregation assay in vitro
Autoaggregation assays were performed according to Del Re (Del Re et al., 2000) with certain modifications. Autoaggregation ability was measured as autoaggregation percentage. The isolate was cultured on MRS for 24 h in an anaerobic jar at 30 ºC, then the cells were fuged for 15 min at 5000 g. The pellet was washed three times in sterile PBS and resuspended in sterile PBS to adjust an OD 600 of 0.25 ± 0.05 (Collado et al., 2008). The autoaggregation assay was done by the bacterial suspension. The bacterial suspension of 4 mL was mixed for 10 s and incubated at room temperature for different times (0 h ,2 h ,4 h ,6 h ,8 h ,10 h, 20 h). The absorbance (A) of bacterial suspension was measured at OD600. The autoaggregation percentage is expressed as: A%=(A0 -At)/A0 x 100, where At represents the absorbance at timet=2,4,6, 8, 10, 20h and A0 represents the absorbance at t=0.
Coaggregation assay in vitro
The method for preparing the cell suspensions for coaggregation was the same as that for autoaggregation assay. The bacterial pathogens used in this study were Vibrio parahaemolyticus (V. parahaemolyticus), Staphylococcus aureus (S.aureus), Shigella, Listeria monocytogenes (L. monocytogenes), Escherichia coli O157 (E.coli O157), Salmonella typhimurium (S.typhimurium), Proteusbacillus vulgaris (P.vulgaris). Then equal volumes (2 mL) of the isolate bacterial suspension and pathogen bacterial suspension were mixed together for 10 s. The absorbance (A) at OD600 of the suspension was measured after 0 h, 2 h, 4 h, 6 h, 8 h, 10 h, 20 h of incubation at room temperature(Kos, 2003). The coaggregation percentage is expressed as: A%=(1-Amix/A0) x 100,where Amix represents the absorbance at timet=2,4,6,8,10, 20 h; A0 represents the absorbance att=0.And each assay was performed in triplicate to correct for intra-assay variation.
Antimicrobial activity assay
The bacterial pathogens, V. parahaemolyticus, Vibrio Harveyi (V. Harveyi), Vibrio alginolyticus (V. alginolyticus), S. aureus, Shigella, E. coli O157, S. typhimurium, P. vulgaris and Bacillus subtilis (B. subtilis), were cultured in LB at 37 ºC for 24 h. L. monocytogenes was cultured in BHI at 37 ºC for 24 h. Lactococcus lactis subsp. cremoris (L. lactis subsp. cremoris) was cultured in MRS for 24 h in an anaerobic jar at 37 ºC. Aspergillus niger (A. niger) was cultured in PDA at 37 ºC for 24 h. The supernatant from cultures of the isolate was prepared by centrifugation at 5000 g for 15 min, sterilized by passage through 0.45 µm Millipore membrane (Millipore, USA) (Balcázar et al., 2008). Antimicrobial activity assay was measured by Oxford Cup. The indicator strains subcultured in culture medium agar plates were flooded with 100 µL of indicator bacteria. The Oxford Cups including the supernatant of 200 µL were placed on the surface of the agar (four Oxford Cup in each plate, one was used for control with no fermentation MRS).Then the plates were incubated in the agar according to their requirements for 24 hours. After the incubation, the diameter of the halos displayed the inhibitory activity.
Result and Discussion
Isolation and identification of the bacterial strain from the fish intestine
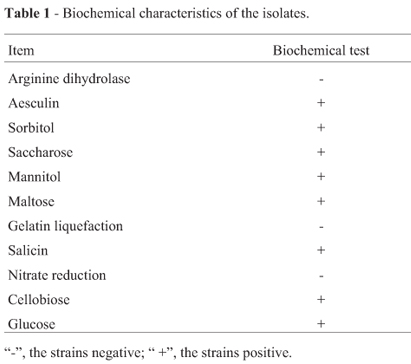
50 strains were isolated from the fish intestines samples, of which 1 isolate strain was considered to be lactic acid bacteria according to the morphological, biochemical characteristics and the lactic acid metabolic products. The isolate strain showed initial characterization of Gram-positive and no spore formation, and could produce small colonies without pigment and grow well in MRS broth at 30 ºC. The strain could produce acid by pH determination after fermentation, gas from glucose and dextran from sucrose, and hydrolyze arginine (Harrigan 1990). The metabolism was consistent with facultative anaerobes. The strain was able to ferment saccharose, mannitol, maltose, cellobiose, glucose and sorbitol. The result of biochemical tests were showed in Table.1. The characteristics of the strain were the same as those of lactic acid bacteria.
PCR amplification of 16S rDNA sequence and phylogenetic tree
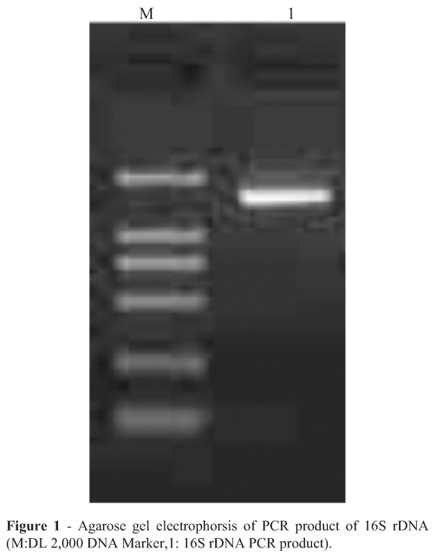
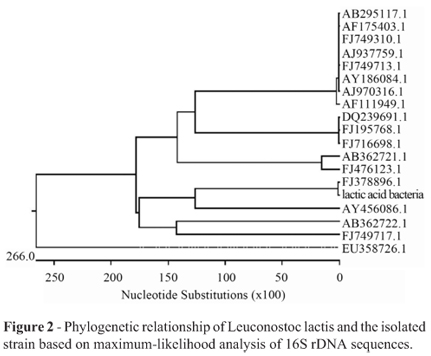
Judgement from the bands of the PCR amplification using two primers in an ethidium stained agarose gel, the length of PCR product was about 1500 bp (Figure 1).The phylogenetic analysis was performed with the 16S rDNA gene sequences of the 19 taxa most closely related to the presumed LAB. The phylogenetic tree based on maximum likelihood was shown in Figure 2. The strain of highest similarity was Leuconostoc citreum strain W-4 (FJ378896.1 in GenBank), and the second highest similarity was Leuconostoc garlicum (AY456086.1 in GenBank). It eventually became clear that the isolate could be assigned to species. Sequencing result established the character of Leuconostoc lactis, so that the isolate in the study was available. Based on 16S rDNA sequence analysis, the isolate above was identified as a strain of lactic acid bacteria, Leuconostoc lactis.
Effect of pH on survival rate of the isolate
pH value was one of factors influencing survival of bacteria in gastric juice, and so the different pH values were investigated about its effect on the survival of bacteria. When pH was 1.5, the number of living bacteria was only 0.43x106 cfu/mL, obviously less than the 6.8x106 cfu/mL living bacteria in control group that pH was 5.5 (Figure 3). The result indicated that pH value could have the key effect on the isolated strain and the viable counts of the isolate came down with pH value decreasing, but the isolate still could survive well in acid condition.
The isolate tolerance for the bile salt
Tolerance to bile is important for the probiotic strains to grow and survive in the digestive tract. In this experiment, the number of living bacteria was showed in Figure 4. Compared to the control (0 g/L), the viable bacteria decreased from 6.3x106 to 1.7x106 cfu/mL when the bile concentration was 4 g/L. The isolate could tolerate bile, but high bile concentration could affect the survival of the isolate.
Survival in Trypsinase and Pepsin in vitro
The survival of the bacterial strain in pepsin solution is showed Figure 5 at pH 3.0. The number of viable bacteria could maintain at 106 cfu/mL. But different pepsin concentrations had an effect on the survival of the bacterial. When the pepsin concentration was 10 g/L, the viable counts of the strain decreased to 1.7x106 cfu/mL, compared to the control (0 g/L) 3.3x106 cfu/mL. According to the result, the isolate could survive well under the action of pepsin, but the pepsin could influence survival of the bacteria to a certain extent.
Tolerance of the isolate to trypsinase was examined in PBS containing trypsinase concentrations from 0 to 7 g/L with incubating 6 h. Compared with the control group (0 g/L), the viable bacteria reached to 106 cfu/mL at the trypsinase concentrations 1, 3, 5, 7 g/L. The result (Figure 6) showed that the isolate could survive better in trypsinase condition. When the trypsinase concentrations increased, the viable counts of the strain decreased. The trypsinase concentration 0 g/L responded to the viable count, 38x106 cfu/mL, and the trypsinase concentration 7 g/L responded to the viable count, only 3.33x106 cfu/mL. In Figure 6, the trypsinase concentrations were 0 to 3 g/L, the number of the viable count decreased rapidly, then tended to steady from 3 to 7 g/L.
If Lactic acid bacteria was used as dietary adjunct, it must tolerate the stomach acidity and survive in the enzyme solution of digestive tract (Conway et al., 1987). The isolate, Leuconostoc lactis, survived well under the action of pepsin and trypsinase in the experiment.
Antibiotic susceptibility of the isolate
Antibiotic resistance is an increasing worldwide problem. The LAB is considered as generally recognized as safe (GRAS) by the FDA (Food and Drug Administration, USA) (Svensson 1999). With the antibiotic being used widely, some of the LAB appeared antimicrobial resistance. Resistance or susceptibility to antibiotic had deserved a special consideration in terms of classification of lactic acid bacteria for its association with human infections (Felten et al., 1999). For the antibiotics assayed, inhibition halos had been compared to results published for NCCLS reference. The isolate was sensitive to common antibiotic used in clinical applications, however represented resistence against cephalothin, ceftriaxone, imipenem and tobramycin (Table.2). The observation indicated that the isolate showed low probability of antibiotic resistance.
Autoaggregation assay
Autoaggregation of probiotic strains appeared to be related to adhesion to intestinal epithelial cells (Boris et al., 1997; Del Re et al., 2000; Reid et al., 1988). The autoaggregation percentage was measured by the sedimentation rate. Result showed that the isolate could autoaggregate itself and the autoaggregation percentage increased from 7.8% to 23.29% during 2-20 h (Figure 7). Compared with other probiotic strains, autoaggregation ability of the isolate was very normal. The percentage of autoaggregation of Bifidobacteria.animalis subsp.lactic BB12 and Lactobacillus casei Shirota were respectively 36.7% and 17.9% (Yong et al., 2010).
Coaggregation assay
Coaggregation abilities may form a barrier that prevents colonization by pathogenic microorganisms (Boris et al., 1997; Del Re et al., 2000; Reid et al., 1988). Coaggregation with pathogens, may constitute an important host defense mechanism against infection (Rickard et al., 2003). In the study, coaggregation of the isolate with seven enteropathogens was examined. Results showed that the isolate coaggregated seven enteropathogens and coaggregation percentage would increase with the prolongation of incubation time. After 20 h of incubation of a mixed suspension (Figure 8), the isolate coaggregated the enteropathogens with the high percentage for V. parahaemolyticus (31.21%) and L. monocytogenes (29.44%), the middle coaggregation percentages for Shigella (24.36%), S.aureus (24.41%) and P. vulgaris (20.99%), and the low coaggregation percentages for E. coli O157 (10.74%) and S.typhimurium (16.49%). The autoaggregation percentage was higher than the coaggregation percentage of seven enteropathogens at 2 h, 4 h, 6 h. But at 20 h, the percentage of coaggregation with V. parahaemolyticus (31.21%), L. monocytogenes (29.44%), Shigella (24.36%) and S.aureus (24.41%) were higher than isolate itself (Figure 8).
The abilities of bacterial autoaggregation and coaggregation were beneficial properties, which were respectively related to cell adherence and inhibitation against pathogen, and played an important role in intestinal tract (Jankovic et al., 2003). The isolate could have the ability of aggregating itself and coaggregating other bacteria, which could be helpful for the healthy of fish in aquaculture.
Antimicrobial activity assay
The ability to inhibition against pathogenic bacteria is used to select potentially probiotic bacteria. In this experiment, the indicator strains contained Gram-positive bacteria, Gram-negative bacteria and Fungi. The supernatant of the isolate didn't showed inhibition to L. monocytogenes, L. lactis subsp.cremoris, A.niger, most effectively inhibition toward E.coli O157, S. typhimurium, B. subtilis, p.vulgaris, V. parahaemolyticus, V. alginolyticus, V. harveyi and Shigella, low inhibition toward S. aureus, as can be seen from Table 3. The metabolite of LAB, such as H2O2, ethanoic acid, lactic acid and lactein were found to inhibit a wide variety of pathogens (Ocana et al., 1999). But it needed to be studied which metabolite played an important role in inhibiting pathogens for the isolate.
Conclusion
In this study, the strain of lactic acid bacteria, Leuconostoc lactis, isolated from marine fish, could be better adaptive to survivorship and colonization in the intestinal tract in accordance with its aggregation ability and tolerance for pH, bile, trypsinase and pepsin conditions. The inhibition effect on the growth of pathogens, the antibiotic little susceptibility and coaggregation assay indicated its potentially probiotic properties. The above result indicated that the isolate, Leuconostoc lactis, could be used as the perspectival strain for probiotics in marine aquaculture.
Submitted: May 25, 2011;
Approved: June 5, 2012.
All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License CC BY-NC.
- Angulo FJ (2000) Antimicrobial agents in aquaculture: Potential impact on health. APUA Newsletter 18:1-6.
- Balcázar JL, Blas ID, Ruiz-Zarzuela I, Cunningham D, Vendrell D, Múzquiz JL (2006) The role of probiotics in aquaculture. Vet Microbiol 114:173-186.
- Balcázar JL, Vendrell D, De Blas I, Ruiz-Zarzuela I, Muzquiz JL, Girones O (2008) Characterization of probiotic properties of lactic acid bacteria isolated from intestinal microbiota of fish. Aquaculture 278:188-191.
- Bauer AWKW, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Tech Bull Regist Med Technol 36:49-52.
- Boris S, Suárez JE, Barbés C (1997) Characterization of the aggregation promoting factor from Lactobacillus gasseri, avaginal isolate. J Appl Microbiol 83:413-420.
- Collado M, Meriluoto J, Salminen S (2008) Adhesion and aggregation properties of probiotic and pathogen strains. Eur Food Res Technol 226:1065-1073.
- Conway PL, Gorbach SL, Goldin BR (1987) Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J Dairy Sci 70:1-12.
- De Man JC, Rogosa M, Sharpe ME (1960) A medium for the cultivation of lactobacilli J Appl Microbiol 23:130-135.
- Del Re B, Sgorbati B, Miglioli M, Palenzona D (2000) Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum Lett Appl Microbiol 31:438-442.
- Felten A, Barreau C, Bizet C, Lagrange PH, Philippon A (1999) Lactobacillus species identification, H2O2 production, and antibiotic resistance and correlation with human clinical status. J Clin Microbiol 37:729-733.
- Harrigan WF (1990) Laboratory Methods in Food and Dairy Microbiology. Academic Press, London.
- Ishida Y, Sugita H (2000) Methods in Microbiology for Marine Environmental Assessment. Koseisha-Koseikaku, Tokyo.
- Jankovic I, Ventura M, Meylan V, Rouvet M, Elli M, Zink R (2003) Contribution of aggregation-promoting factor to maintenance of cell shape in Lactobacillusgasseri 4B2. J Bacteriol 185:3288-3296.
- Kos B, SuSkovic J, Vukovic S, Simpraga M, Frece J, MatoSic S (2003) Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol 94:981-987.
- Ocana VS, Pesce De Ruiz Holgado AA, Nader-Macias ME (1999) Characterization of a bacteriocin-like substance produced by a vaginal Lactobacillus salivarius strain. Appl Environ Microbiol 65:5631-5635.
- Reid G, McGroarty JA, Angotti R, Cook RL (1988) Lactobacillus inhibitor production against Escherichia coli and coaggregation ability with uropathogens. Can J Microbiol 34:344-351.
- Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, Roberfroid M, Rowland I, Cherbut C, Klaenhammer TR (2003) New scientific paradigms for probiotics and prebiotics. J Cl Gastroenterol 37:105-118.
- Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS (2003) Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol 11:94-100.
- Ringø E, Gatesoupe FJ (1998) Lactic acid bacteria in fish: A review. Aquaculture 160:177-203.
- Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406-425.
- Stoesser G, Baker W, Van Den Broek A, Camon E, Garcia-Pastor M, Kanz C, Kulikova T, Lombard V, Lopez R, Parkinson H, Redaschi N, Sterk P, Stoehr P, Tuli MA (2001) The EMBL nucleotide sequence database. Nucleic Acids Res 29:17-21.
- Svensson U (1999) Probiotics: A Critical Review. Horizon Scientific Press, UK.
- Verschuere L, Rombaut G, Sorgeloos P, Verstraete W (2000) Probiotic bacteria as biological control agents in aquaculture. Microbiol Mol Biol Rev 64:655-671.
- Yong L, Yong Z, Yan B, Heping Z (2010) Study on cell surface properties and inhibitory effects on pathogens of four probiotic strains. Journal of Chinese Institute of Food Science and Technology. 10:28-34.
Send correspondence to
Publication Dates
-
Publication in this collection
15 Nov 2013 -
Date of issue
Sept 2013
History
-
Received
25 May 2011 -
Accepted
05 June 2012