Abstract
The aims of this study were to standard and optimize a qPCR protocol with FAM-BHQ1 probe, and to compare its sensitivity against TaqMan qPCR and PCR methods to diagnose shrimp WSD. The FAM-BHQ1 qPCR presented higher clinical sensitivity and showed to be a robust alternative to detect WSSV in clinical samples.
clinical samples; sensitivity; real-time PCR; WSSV; WSD
Comparative analysis of conventional PCR and real-time PCR to diagnose shrimp WSD
C.A.G. LealI; G.A. Carvalho-CastroI; A.C. CottorelloII; R.C. LeiteI; H.C.P. FigueiredoI, *
ILaboratório de Doenças de Animais Aquáticos, Escola de Veterinária, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brazil
IILaboratório Nacional Agropecuário, Ministério da Agricultura, Pedro Leopoldo, MG, Brazil
Send correspondence to Send correspondence to H.C.P. Figueiredo AQUACEN-Laboratory of Aquatic Animal Diseases Av. Antônio Carlos 6627, Pampulha Belo Horizonte, MG, Brazil E-mail: henrique@dmv.ufla.br
ABSTRACT
The aims of this study were to standard and optimize a qPCR protocol with FAM-BHQ1 probe, and to compare its sensitivity against TaqMan qPCR and PCR methods to diagnose shrimp WSD. The FAM-BHQ1 qPCR presented higher clinical sensitivity and showed to be a robust alternative to detect WSSV in clinical samples.
Key words: clinical samples, sensitivity, real-time PCR, WSSV, WSD.
White spot disease (WSD) is one of the most important viral disease for marine shrimp culture, causing serious mortalities to economically important species, such as Penaeus monodon, Litopenaeus vannamei and L. stylirostris (Sanchéz-Martinéz et al., 2007). Etiologic agent of WSD, the white spot syndrome virus (WSSV) is a bacilliform, non-occluded enveloped, double-stranded DNA virus of Nimaviridae family (Sritunyalucksana et al., 2006; Escobedo-Bonilla et al., 2008). The illness was firstly reported in 1992 in Taiwan. Since then, the continuous spread of disease has been described, mainly throughout the regions of important shrimp production (Asia and the Americas) (Sritunyalucksana et al., 2006; Escobedo-Bonilla et al., 2008; Nunan and Lightner, 2011). Currently, WSD is considered one of the main barriers for successful expansion of shrimp industry worldwide (Nunan and Lightner, 2011; OIE, 1997).
Several methods have been used to diagnose WSD in shrimp, including histopathology, immunological assays, in situ hybridization and molecular techniques (Durand and Lightner, 2002; Chou et al., 2011; Samanman et al., 2011). PCR-based assays offer high degrees of sensitivity and specificity (Chou et al., 2011); being the Nested PCR protocol developed by Lo et al. (1996), the gold standard recommended by the OIE Manual of Diagnostic Tests for Aquatic Animals (OIE, 2009). Despite of the advantages, the WSSV Nested-PCR is a two steps, time consuming method and false positive results can be obtained (Nunan and Lightner, 2011). Quantitative Real-time PCR (qPCR) has been successfully used to quantify different viral infections in shrimp, showing to be fast, sensitive and specific (Jang et al., 2009). Many different Sybr-green and TaqMan qPCR have been developed to detect WSSV (Durand and Lightner, 2002; Sritunyalucksana et al., 2006; Jang et al., 2009; Chou et al., 2011). However, these techniques were not included in the OIE Manual (OIE, 2009) as a recommended method. Contradictory results, variable reproducibility and sensibility data have been verified while using that (Durand and Lightner, 2002; Durand et al., 2003; Sritunyalucksana et al., 2006).
The aim of this study was to standard a qPCR protocol based in dual-labeled hydrolysis probe technology, with non-fluorescent quencher (Black Hole Quencher, BHQ1), to diagnose WSD. In addition, to compare the clinical sensitivity of the this protocol against previously developed PCR (Nested PCR and TaqMan qPCR) to detect WSSV in clinical samples of diseased shrimp.
Diseased and health Litopenaeus vannamei were collected during outbreaks of WSD in shrimp farms, located in two Brazilian States (Santa Catarina and Bahia). The samples were 96% ethanol preserved and immediately conducted to LANAGRO-National Animal and Plant Laboratory of Brazilian Ministry of Agriculture, situated in Pedro Leopoldo (Minas Gerais State). The total shrimp DNA was extracted from three left abdominal pleopods of each animal, using the commercial kits Wizard® DNA Genomic Purification (Promega, USA), DNeasy (Qiagen, USA), and TRIzol (Invitrogen, USA). The amount of extracted DNA was quantified spectrophotometrically with GE NanoVue® Spectrophotometer (GE Healthcare, UK). The DNA samples were stocked at -20 ºC until use.
The primers sets 146F1 (5'ACTACTAACTTCAG CCTATCTAG 3') / 146R1 (5' TAATGCGGGTGTAAT GTTCTTACGA3'), and 146F1/146R1 plus 146F2 (5' GTAACTGCCCCTTCCATCTCCA 3')/146R2 (5' TACGGCAGCTGCTGCACCTTGT 3') were respectively used for one-step and two steps (Nested) WSD PCR. The different qPCR methods were performed using the primer set WSS1011F (5' TGGTCCCGTCCTCATCTCAG 3') / WSS1079R (5'GCTGCCTTGCCGGAAATTA 3') and probe (5' AGCCATGAAGAATGCCGTCTATCACACA 3') (Durand and Lightner, 2002). The primers and probe target the genomic fragment saltI of WSSV (Genbank accession number U50923). Two probe types were tested: a TaqMan probe (Applied Biosystems, USA) labeled with fluorescent dyes 5-carboxyfluoroscein (FAM) on the 5' end and N,N,N',N'-tetramethyl-6-carboxyrhodamine (TAMRA) on the 3' end; and dual-labeled hydrolysis probe with fluorescent dye FAM on the 5' end and the non-fluorescent quencher, Black Hole Quencher® 1 (Sigma-Aldrich, USA), on the 3' end (FAM-BHQ1). All primers used were acquired from Integrated DNA Technologies (IDT, USA).
The one step and two steps WSD PCR were performed as described previously (OIE, 2009). Briefly, PCR mixture of both reactions consisted of: 1X GoTaq Flexi Buffer (Promega); 1.5 mM MgCl2; 200 uM of each dNTP; 100 pmol of each primer; and2Uof GoTaq DNA Polymerase (Promega). 10 uL of one step reaction was used as template for the two steps PCR. Cycling conditions for both assays was 94 ºC for 4 min followed by 39 cycles of 94 ºC for 1 min, 55 ºC for 1 min and 72 ºC for 2 min and a final extension step at 72 ºC for 5 min. The products were visualized in 1% agarose gel electrophoresis containing 0.5 ug.mL-1 of ethidium bromide. The expected amplicon sizes were respectively 1447 and 941 bp, for one and two step reaction. A purified plasmid containing the WSSV genomic sequence U50923 was used as positive control. It was serially 10-fold diluted to standard curve construction in qPCR analysis. Six plasmid dilutions were used to determine the analytical sensitivity of different assays. The amount of plasmid copies ranged from 2 to2x105.
qPCR reactions with TaqMan and FAM-BHQ1 were optimized for primers and probe concentrations, and compared with previously described reaction (Durand and Lightner, 2002). A set of standard curves was created using six dilutions of control plasmid. The primers were evaluated at concentrations ranging from 5 to 100 pmol and probes varied from 5 to 75 pmol per reaction. The standard curves were evaluated in triplicate for each qPCR mixture tested. The best reaction was determined based in: slope factor (-3.1035 to -3.7762); correlation coefficient (above 0.99); and lower average quantification cycle (Cq) for each dilution (Ishi et al., 2007; Bustin et al., 2009). All qPCR assays were performed in an ABI 7500 Real-time System (Applied Biosystems) and TaqMan Universal Master Mix with ROX as passive reference (Applied Biosystems) was used. The qPCR cycling consisted of 95 ºC for 10 min followed by 40 cycles of 95 ºC for 15 seconds and 60 ºC for 60 seconds. Data acquisition and analysis were performed using 7500 Software Version 2.0 (Applied Biosystems). The qPCR data were evaluated and are presented according to MIQE guidelines (Bustin et al., 2009; Bustin, 2010).
The clinical sensitivity of the qPCR protocols were addressed in comparison with conventional PCR methods. Thus, total DNA of 23 clinical samples, 15 WSD positive and 8 WSD negative, were tested using the different techniques and the sensitivity calculated. The effect of different concentrations of template DNA in the clinical sensitivity of conventional and real-time techniques was also determined. Five DNA concentrations (100, 150, 300 and 600 ng per reaction) were tested. Additionally, the clinical sensitivity of qPCR with TaqMan and FAM-BHQ1 were evaluated under low template DNA amounts per reaction (10, 50, 100 and 150 ng). All analyses were performed in triplicate.
Fisher's Exact Test was applied to determine statistical differences among sensitivity of distinct diagnostic methods evaluated. All analysis was performed using SAS Statistical Software STAT Version 6.12 (SAS Institute Inc., USA). A p value of 0.05 or less was considered statistically significant.
Similar optimization results for qPCR with different probes were obtained. Best results were verified with primers and probe concentrations of 90 and 25 pmol, respectively. Optimized TaqMan and FAM-BHQ1 reactions promoted higher correlation coefficient (1.0) for standard curves and better slope factor (-3.45 and -3.42) than previously described TaqMan qPCR (0.99 and -3.19) (Durand and Lightner, 2002). In addition, these reactions generated lower Cq standard deviation (SD) among dilution replicas (data not show). The analytical sensitivity of different methods were 208 plasmid copies for one step PCR, and approximately 2 copies for two step PCR and optimized qPCR techniques. For standard curves, there were no differences in analytical sensitivity between qPCR reactions with distinct probes (TaqMan or FAM-BHQ1).
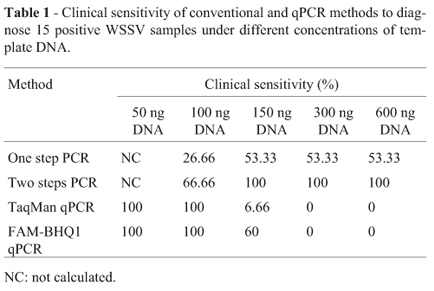
Clinical sensitivity of different methods applied was roughly affected by the template DNA concentrations used. The results for 15 positive WSSV samples evaluated are presented in Table 1. All methods showed high specificity, with only one false positive result for optimized TaqMan qPCR. Conventional PCR methods showed large intrasample variation in the diagnostic results when low amount of template DNA (100 ng per reaction or less) was used. Thus, the clinical sensitivity for 50 ng reactions could not be calculated. qPCR assays with 50 and 100 ng of template DNA showed to be statistically superior to one step (p < 0.0001) and two steps PCR (p = 0.021). In contrast, high DNA concentrations (300 and 600 ng) generated better sensitive results for one (p = 0.001) and two steps PCR (p < 0.0001) compared with real-time methods. Positivity of 100% was obtained with two steps PCR using 300 and 600 ng of template DNA. The presence of high amount of DNA promoted an inhibitory effect in qPCR reactions with TaqMan and FAM-BHQ1. The results were not affected by different methods applied to DNA extraction. The qPCR assays presented negative results for all 15 positive WSD strains, when template DNA concentrations overcame 150 ng per reaction. For this concentration (150 ng), two steps PCR showed the best result (p = 0.008) and no statistical difference were observed between one step PCR and FAM-BHQ1 qPCR (p = 0.5).
The performance of different probes (TaqMan and FAM-BHQ1) in diagnostic qPCR assays was addressed. Significant differences were observed in lowest (10 ng DNA) and highest (150 ng) DNA concentrations tested. FAM-BHQ1 qPCR presented sensitivity of 100% (p < 0.0001) and 60% (p = 0.003) for reactions with 10 ng and 150 ng of template DNA, respectively. Moreover, they were higher and statistically significant compared to optimized TaqMan (60% and 6.66%) results. There was no significant difference (p > 1.00) between the performance of optimized TaqMan and FAM-BHQ1 in 50 and 100 ng DNA reactions (100% sensitivity). However, lower averages Cq for the major of tested samples were obtained with FAM-BHQ1 reaction. That assay did not present any false positive as verified for optimized TaqMan.
The present study was able to standard and to optimize a FAM-BHQ1 qPCR protocol, and to compare its performance with conventional and real-time PCR methods to diagnose WSSV in clinical samples. The analytical sensitivity of two steps PCR and qPCR techniques was similar to described early (Lo et al., 1996; Durand and Lightner, 2002; Sritunyalucksana et al., 2006; Nunan and Lightner, 2011). In contrast, the one step PCR presented contradictory to the parameter reported in OIE Manual (OIE, 2009). Our data for one sep PCR is in accordance with previous publications of Sritunyalucksana et al. (2006) and Nunan and Lightner (2011). Since that reaction was recommended in 1990, PCR technologies and reagents have been submitted to a continuous process of quality improvements (Nunan and Lightner, 2011). Therefore, those advances might allow better results with same protocols, achieved with the use of improved reagents, as verified here and previously (Sritunyalucksana et al., 2006; Nunan and Lightner, 2011).
The clinical sensitivity of distinct PCR techniques was highly affected by template DNA amount used in the reactions. Two steps PCR presented best sensitivity results for reactions with higher concentrations of shrimp DNA (> 150 ng). In contrast, FAM-BHQ1 qPCR showed best performance with low DNA concentrations (< 150 ng). Those reactions presented the same analytical sensitivity, being able to detect plasmid 2 copies. However, for two steps PCR and samples with low target concentration (positive in FAM-BHQ1 reactions), the total shrimp DNA decreases its sensitivity. In spite of the recommended DNA concentration range from 100 ng to 300 ng for two steps PCR (OIE, 2009), an abrupt reduction in sensibility was verified when 100 ng were used. Lo et al. (1996) described 100 ng of DNA as the optimal template concentration for that assay, but, the methodology used to determine that was not presented. Thus, the information accuracy of that cannot be evaluated. Herein, 100% of sensitivity was obtained with DNA levels of 150, 300 and 600 ng, suggesting that the recommended range of template concentration in two steps PCR is slightly higher at minimum level (150 ng). In addition, the DNA amount can securely overcome the maximum level of 300 ng (until 600 ng) without sensitivity loss; however, unspecific amplicons can be verified. High DNA concentrations (> 150 ng) promoted an inhibitory effect in real-time PCR protocols. This phenomenon has been already described by qPCR reactions with total template DNA of other marine invertebrates. The potential inhibitors can be extracted together with nucleic acids, as well as, high concentrations of DNA of those animals can directly inhibit real-time PCR reactions, in a concentration-dependent way (Pan et al., 2008).
TaqMan and FAM-BHQ1 qPCR protocols presented higher clinical sensitivity when 50 and 100 ng of total shrimp DNA was used. These could be valuable tools, mainly to diagnose WSD in larvae and postlarvae shrimp. Those specimens usually present low viral loads and the total amount of DNA obtained can be reduced (Jang et al., 2009); that combination increases the possibility of false negative results, when conventional PCR techniques are applied. Therefore, qPCR protocols can solve this problem, allowing to detect WSSV in young animals; the main kinds of shrimp transported between the farms.
Different performances were observed in qPCR sensitivity with TaqMan and FAM-BHQ1 probes for WSSV detection. TaqMan are dual-labeled hydrolysis probes constituted of the fluorophore FAM in 5' extremity and the fluorescent quencher TAMRA at 3' (Daum et al., 2004). TAMRA fluoresces; whereas Black Hole Quencher 1 (BHQ1) is a dark quencher, which re-emits its energy as heat rather than light (Yang et al., 2009). The background fluorescence of TAMRA may reduce the sensitivity of qPCR assays (Daum et al., 2004; Yang et al., 2009). This behavior was verified for the tested WSSV qPCR reactions. TaqMan assay had overall worst results than reactions using FAM-BHQ1 probes. FAM-BHQ1 qPCR promoted lower intra-assay variation and Cq for tested samples. Similar results were found by Yang et al. (2009) evaluating the performance of TaqMan and BHQ1 quenched probes to detect viral and bacterial respiratory pathogens. FAM-BHQ1 qPCR for WSSV detection showed a constant 100% clinical sensitivity in reactions within the recommended range of DNA concentrations (10-100 ng per reaction). In contrast, TaqMan qPCR presented a significant sensitivity decrease in reactions with low DNA amount (10 ng). Those data clarify the advantages of standardized FAM-BHQ1 qPCR in comparison to TaqMan assay.
In conclusion, the standardized FAM-BHQ1 qPCR protocol showed to be a fast, robust and viable tool to diagnose WSSV in clinical samples. It could be used as an alternative to two steps PCR, mainly to diagnose WSD in larvae and postlarvae shrimp.
Acknowledgments
This work was supported by CNPq Grant 578767/2008-2, Ministry of Fisheries and Aquacultures and by National Institute for Science and Technology INCT-573899/2008-8 INCT/CNPq/UFMG. Carlos A. G. Leal fellowship was provided by CNPq (Grant 158671/2010-4).
Submitted: September 9, 2011; Approved: September 10, 2012.
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem 55:611-622.
- Bustin SA (2010) Why the need for qPCR publication guidelines? The case for MIQE. Methods 50:217-226.
- Chou P, Lin Y, Teng P, Chen P, Lee P (2011) Real-time target-specific detection of loop-mediated isothermal amplification for white spot syndrome virus using fluorescence energy transfer-based. J Virol Methods 173:67-74.
- Daum LT, Ye K, Chambers JP, Santiago J, Hickman JR, Barnes WJ, Kruzelock RP, Atchley DH (2004) Comparison of TaqMan and Epoch Dark Quenchers during real-time reverse transcription PCR. Mol Cellular Probes 18:207-209.
- Durand SV, Lightner DV (2002) Quantitative real time PCR for the measurement of white spot syndrome virus in shrimp. J Fish Dis 25:381-394.
- Durand SV, Redman RM, Mohney LL, Tang-Nelson K, Bonami JR, Lightner DV (2003) Qualitative and quantitative studies on the relative virus load of tails and heads of shrimp acutely infected with WSSD. Aquaculture 213: 9-18.
- Escobedo-Bonilla CM, Alday-Sanz V, Wille M, Sorgeloos P, Pensaert MB, Nauwynck HJ (2008) A review of the morphology, molecular characterisation, morphogenesis and pathogenesis of white spot syndrome virus. J Fish Dis 3:11-18.
- Ishii T, Sootome H, Shan L, Yamashita K (2007) Validation of universal conditions for duplex quantitative reverse transcription polymerase chain reaction assays. Anal Bioch 362:201-212.
- Jang I, Meng X, Seo H, Cho Y, Kim B, Ayyaru G, Kim J (2009) A TaqMan real-time PCR assay for quantifying white spot syndrome virus (WSSV) infections in wild broodstock and hatchery-reared postlarvae of fleshy shrimp, Fenneropenaeus chinensis Aquaculture 287:40-45.
- Lo CF, Ho CH, Peng SE, Chen CH, Hsu HC, Chiu YL, Chang CF, Liu KF, Su MS, Wang CH, Kou GH (1996)White spot syndrome baculovirus (WSBV) detected in cultured and captured shrimp, crab and other arthropods. Dis Aquat Org 27:215-225.
- Nunan LM, Lightner DV (2011) Optimized PCR assay for detection of white spot syndrome virus (WSSV). J Virol Methods 171:318-321.
- Office International des Epizooties-OIE (1997) .Aquatic Animal Health Code, 1997 Edition. OIE, Paris.
- Office International des Epizooties-OIE (2009) Diagnostic Manual for Aquatic Animal Diseases, 6th edition. OIE, Paris.
- Pan M, McBeath AJA, Hay SJ, Pierce GH, Cunningham CO (2008) Real-time PCR assay for detection and relative quantification of Liocarcinus depurator larvae from plankton samples. Mar Bio 153:859-870.
- Samanman S, Kanatharana P, Chotigeat W, Deachamag P, Thavarungkul P (2011) Highly sensitive capacitive biosensor for detecting white spot syndrome virus in shrimp pond water. J Virol Methods 173:75-84.
- Sanchéz-Martinéz JC, Aguirre-Guzmán G, Mejía-Ruíz H (2007) White Spot Syndrome Virus in cultured shrimp: A review. Aquaculture Res 122:1-15.
- Sritunyalucksana K, Srisala J, McColl K, Nielsen L, Flegel TW (2006) Comparison of PCR testing methods for white spot syndrome virus (WSSV) infections in penaeid shrimp. Aquaculture 255: 95-104.
- Yang GP, Erdman DD, Tondella ML, Fields BS (2009) Evaluation of tetramethylrhodamine and black hole quencher 1 labeled probes and five commercial amplification mixes in TaqMan® real-time RT-PCR assays for respiratory pathogens. J Virol Methods 169:288-290.
Send correspondence to
Publication Dates
-
Publication in this collection
15 Nov 2013 -
Date of issue
Sept 2013
History
-
Received
09 Sept 2011 -
Accepted
10 Sept 2012