Abstract
A previously reported o-nitrobenzaldehyde (ONBA) degrading bacterium Pseudomonas sp. ONBA-17 was further identified and characterized. Based on results of DNA base composition and DNA-DNA hybridization, the strain was identified as P. putida. Its degradation effect enhanced with increase of inoculum amount and no lag phase was observed. Higher removal rate was achieved under shaking conditions. All tested ONBA with different initial concentrations could be completely degraded within 5 d. In addition, degradative enzyme(s) involved was confirmed as intra-cellular distributed and constitutively expressed. Effects of different compounds on relative activity of degradative enzyme(s) within cell-free extract were also evaluated. Finally, 2-nitrobenzoic acid and 2, 3-dihydroxybenzoic acid were detected as metabolites of ONBA degradation by P. putida ONBA-17, and relevant metabolic pathway was preliminary proposed. This study might help with future research in better understanding of nitroaromatics biodegradation.
Pseudomonas putida; biodegradation; metabolic pathway; nitroaromatics
RESEARCH PAPER
Further characterization of o-nitrobenzaldehyde degrading bacterium Pseudomonas sp. ONBA-17 and deduction on its metabolic pathway
Fang-Bo YuI; Xiao-Dan LiII; Shinawar Waseem AliIII; Sheng-Dao ShanI; Lin-Ping LuoI; Li-Bo GuanI
IDepartment of Environmental Sciences, Zhejiang Agricultural and Forestry University, Linan, China
IIInstitute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, China
IIIInstitute of Agricultural Sciences, University of the Punjab, Lahore, Pakistan
Correspondence Correspondence: F.B. Yu College of Environment and Resource Sciences Zhejiang Agricultural and Forestry University 88 Huanchengbei Road 311300 Linan, China E-mail: yufangbo@aliyun.com
ABSTRACT
A previously reported o-nitrobenzaldehyde (ONBA) degrading bacterium Pseudomonas sp. ONBA-17 was further identified and characterized. Based on results of DNA base composition and DNA-DNA hybridization, the strain was identified as P. putida. Its degradation effect enhanced with increase of inoculum amount and no lag phase was observed. Higher removal rate was achieved under shaking conditions. All tested ONBA with different initial concentrations could be completely degraded within 5 d. In addition, degradative enzyme(s) involved was confirmed as intra-cellular distributed and constitutively expressed. Effects of different compounds on relative activity of degradative enzyme(s) within cell-free extract were also evaluated. Finally, 2-nitrobenzoic acid and 2, 3-dihydroxybenzoic acid were detected as metabolites of ONBA degradation by P. putida ONBA-17, and relevant metabolic pathway was preliminary proposed. This study might help with future research in better understanding of nitroaromatics biodegradation.
Key words: Pseudomonas putida, biodegradation, metabolic pathway, nitroaromatics.
Introduction
Nitroaromatic compounds occur as intermediates or by-products in a number of industrial processes. Due to improper storage, use and disposal, nitroaromatic compounds have been released into environment which resulted in their emergence as environmental contaminants (Lee et al., 2005; Jeon and Madsen, 2013). These compounds are toxic and recalcitrant to degradation due to stability of nitroaromatic ring structure. Microbial metabolism plays a very important role in the degradation or detoxification of such chemicals in the environment. Successful removal by implanted bacteria has been previously reported for many compounds (Perelo, 2010; Ramos et al., 2011; Chakraborty et al., 2012).
O-nitrobenzaldehyde (ONBA) is an important intermediate for the synthesis of a number of chemicals (Liu et al., 2012). In China, annually, a large quantity of wastewater containing ONBA is generated and discharged into environment, causing severe pollution hazards. Thus, it is essential to treat such wastewater prior to its discharge even though no acceptable limit of ONBA has been specifically set.
Previously, an aerobic ONBA degrading bacterium, designated as ONBA-17, was isolated from activated sludge and identified as Pseudomonas sp. (Yu et al., 2006). Subsequently, the feasibility of using ONBA-17 chromosomally marked with gfp gene to bioaugment a sequencing batch reactor treating ONBA synthetic wastewater was confirmed (Yu et al., 2010). However, further characterization of the bacterium is still needed. In this study, we described further identification and degradation characteristics of ONBA-17. Distribution and expression of degradative enzyme(s) in cell-free extract were also investigated. Besides, ONBA metabolic pathway within this bacterium was preliminary deduced.
Materials and Methods
Chemicals
ONBA, 2-nitrobenzoic acid and 2, 3-dihydroxybenzoic acid were purchased from Sigma Aldrich (Beijing, China). Other chemicals and reagents were of the highest grade and employed without further purification.
Bacterial strains and medium
Bacterial strains used in this study but ONBA-17 were provided by Dr. Lei Ma, Nanjing Agricultural University. Compositions of minimal salts medium (MSM) were as follows: MgSO4 0.25 g, NH4NO3 1 g, KH2PO4 2 g, K2HPO4 7.5 g, NaCl 1 g, double distilled water 1 L, pH 7.6.
DNA base composition and DNA-DNA hybridization
G+C content of genomic DNA was determined by thermal denaturation method (Marmur and Doty, 1962), using Escherichia coli K-12 as a reference. Genomic DNA from ONBA-17, P. monteilii ATCC 700476, P. plecoglossicida ATCC 700383 and P. putida KT2440 (ATCC 47054) were extracted and purified according to standard procedures (Sambrook and Russell, 2001). DNA-DNA hybridization was carried out according to the method of Huss et al. (1983) using a Perkin Elmer Lambda 35 UV/VIS spectrometer equipped with a PTP-6 Peltier system.
Degradation in MSM by ONBA-17
The effect of inoculum amount (OD600 0.1-0.6, 30 °C, pH 7.6, 160 rpm) on ONBA degradation was examined according to Yu et al. (2013). The washed cells were inoculated into the MSM containing 100 mg/L of ONBA. Cultivation was conducted in a rotary shaker (160 rpm) for 72 h and residual ONBA was determined by gas chromatography (GC) at 8-h intervals according to the method described by Yu et al. (2006). The analysis was conducted with a Shimadzu gas chromatograph (GC-14B) equipped with a FID (flame ionization detector). Moreover, effects of incubation conditions (shaking vs. static) on biodegradation were examined according to the method described by Hussain et al. (2007). Biodegradation assay (OD600 0.4, 30 °C, pH 7.6, 160 rpm), containing different initial concentrations of ONBA (50-300 mg/L), were also conducted. All above experiments were performed in quadruplicate.
Removal of ONBA by cell-free extract
Cell-free extract was prepared according to the method used in Yu et al. (2013). The assay to quantify removal of ONBA by cell-free extract was performed in 0.05 M phosphate buffer (pH 7.6). Each reaction vial comprised 20 µL of the cell-free extract prepared as described above in 2 mL of 0.05 M phosphate buffer (pH 7.6) containing 50 mg/L of the test substrate. At regular intervals, the reaction was stopped by adding 0.1 M NaOH. The remaining ONBA concentration in the vial was determined as described in Yu et al. (2006). Total protein was estimated by the method of Lowry et al. (1951), using bovine serum albumin (Sigma, Beijing, China) as the standard.
Localization of degrading enzyme(s) was conducted by the method of osmotic shock (Harold and Leon, 1965). Solutions were each transferred (10%, v/v) into phosphate buffer solution (0.05 M, pH 7.4) containing 20 mg ONBA per liter and incubated at 30 °C with shaking at 120 rpm. The reaction mixtures at zero time and 2 h were examined against a blank control containing the same reaction mixture, except that ONBA was omitted.
To determine whether enzyme(s) responsible for ONBA biodegradation was inducible or constitutive, the method described by Yu et al. (2008) was used. Cells were grown in the MSM containing 0.5% glucose in the presence or absence of 50 mg/L of ONBA. The reaction mixture (1 mL) contained phosphate buffer (0.05 M, pH 7.4), ONBA (50 mg/L) and cell crude extract. Reactions were performed at 30 °C for 1 h without shaking, and the residual ONBA was quantified as mentioned above. The pH and temperature ranges of degradative enzyme(s) within the extract were determined as described by Liang et al. (2005). The effects of potential inhibitors or activators on the enzyme(s) were determined by addition of various metal salts (LiCl, AgNO3, MgCl2, CuCl2, ZnCl2, CdCl2, BaCl2, MnCl2, CaCl2, NiCl2, CoCl2, FeCl2 and FeCl3), surfactants (SDS, Tween 20 and Tween 80), and chelating agent (EDTA) into the reaction mixture (5 mL) containing phosphate buffer (0.05 M, pH 7.4), ONBA (100 mg/L) and 100 µL cell-free extract. Final concentration of above test substances was 0.2 mM. Subsequently, reactions were performed at 30 °C for 12.5 min without shaking, and the residual ONBA was quantified as above mentioned.
Deduction of ONBA degrading pathway
50 mL Luria-Bertani (LB)-grown cells were harvested and inoculated into 50 mL of MSM supplemented with 100 mg/L of ONBA and incubated at 30 °C for 48 h. Samples taken at 0, 2, 6, 12, 24, 36 and 48 h were immediately treated and re-dissolved in acetonitrile as described by Yu et al. (2006). Trimethylsilyl derivatives were prepared with N, O-bis(trimethylsilyl)-trifluoroacetamide according to the method provided by the distributor (Alltech Associates Inc.). Gas chromatopgraphy-mass spectrometry (GC-MS) analysis was conducted with TRACE GC-DSQ (Thermo Finnigan) equipped with a DB-5MS capillary column (25 m x 0.25 mm i.d. x 0.25 m). Chromatography program was as follows: initial column temperature of 160º C, isothermal for 1 min, temperature increase of 20 ºC/min to 280 °C, and isothermal for 3 min. The electron impact mass spectra were obtained at 70 eV and monitored in a range of 50 to 400 m/z. Product identities were confirmed by comparison of retention times (Rt) and MS fragmentation profiles to authentic chemical standards. Catechol 1, 2-dioxygenase, catechol 2, 3-dioxygenase, nitrite and ammonia concentrations assays were performed as previously described (Haigler and Spain, 1993). The experiments were performed in triplicate.
Results and Discussion
Further identification of strain ONBA-17
Results show that DNA G+C contents of strain ONBA-17, ATCC 700476, ATCC 700383 and KT2440 were 61.4 ± 0.2 mol%, 62.8 ± 0.2 mol%, 60.5 ± 0.3 mol% and 61.7 ± 0.1 mol%, respectively, close to those of other species of the genus Pseudomonas (Nishimori et al., 2000). DNA-DNA relatedness value between strain ONBA-17 and P. putida KT2440 was 85.3 ± 0.4%, which is above the value of approximately 70% that has been suggested as a threshold to delineate bacterial species (Grimont, 1999). The values between ONBA-17 and the other two strains were 46.0 ± 0.2% (ATCC 700383) and 35.4 ± 0.1% (ATCC 700476), respectively. Thus, based on the results of morphological, physiological and biochemical characterization, phylogenetic analysis of 16S rDNA gene sequence (Yu et al., 2006), DNA G+C content measurement and DNA-DNA hybridization, strain ONBA-17 was identified as P. putida.
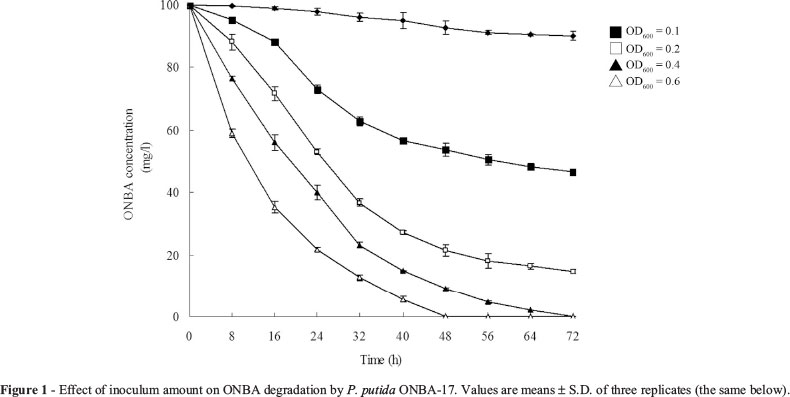
Effect of inoculum amount on ONBA degradation
As shown in Figure 1, degradation effect enhanced with increase of inoculum amount. In cultures inoculated with the highest initial cell density (OD600 = 0.6), ONBA degradation initiated rapidly within 8 h, apparently there was no lag phase and more than 90% of the test substrate was degraded within 40 h. Similarly, in cultures inoculated with lower initial cell density (OD600 = 0.4), the same result was observed and 100 mg/L ONBA could be thoroughly degraded after 3 days of incubation. However, the percentage of ONBA removal decreased sharply and complete degradation occurred in longer time when cell density lower than 0.4. The percentages of ONBA removal at OD600 0.2 and 0.1 were 85.3% and 53.3%, respectively, after 72 h of incubation.
Previous studies reported that the degrading efficiency of recalcitrant compound was dependent on initial inoculum amount (Xu et al., 2011). In cultures with low inoculum densities, there were long lag periods before efficient degradation started as reported by Anwar et al. (2009). When low inoculum amount was used, only a part of introduced cells could survive from the initial competition and take part in degradation. Furthermore, high inoculum amount could partly compensate for the initial population decline (Anwar et al., 2009). However, our results are not in accordance with above studies. Although ONBA-degrading efficiency was enhanced by increasing inoculum amount, there was no lag phase observed throughout the test. Similarly, the phenomenon has also been observed by Chen et al. (2011) and Yu et al. (2013). The results suggest that strain ONBA-17 could degrade ONBA rapidly even at low inoculum density, which might signify the bacterium was robust and proficient for bioremediation of ONBA-contaminated sites.
A significant difference between biodegradation of ONBA by the strain under static and shaking conditions was noted. Maximum biodegradation of ONBA, up to 100%, was recorded under shaking conditions (30 °C, 160 rpm, ODinitial = 0.4, 72 h). However, the counterpart was only 37.7% as found under static conditions. Better bioavailability of the test substrate to microorganisms coupled with physiochemical degradation might explain the phenomenon to some extent. Besides, non-biological degradation could not be neglected and was more obvious under shaking conditions (data not shown), which implied that aerobic conditions are relatively more conducive for abiotic degradation.
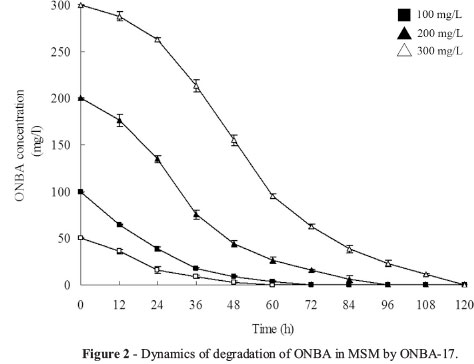
Biodegradation of ONBA with different initial concentrations
Dynamic curves of ONBA degradation with different initial substrate concentrations were presented in Figure 2. At the concentration of 50 and 100 mg/L, ONBA degraded completely within 72 h. At the highest test concentration (300 mg/L), only 79.02% was achieved within 3 d, and complete degradation occurred in a longer time (about 120 h). The degradation rate and concentration were in proportion. Furthermore, P. putida ONBA-17 had not been saturated by the substrate, and increasing concentration could raise the degradation rate.
Removal of ONBA by cell-free extract
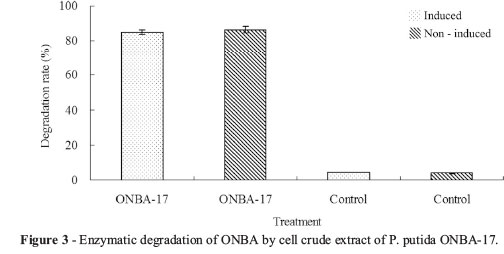
When 20 µL of cell extract prepared was added to 2 mL of reaction buffer containing 50 mg/L ONBA the substrate was completely depleted after 20 h incubation. Under the same conditions, no substrate depletion was recorded while boiled extract was added to the reaction buffer. This indicates that ONBA was transformed by soluble enzymes from cell-free extract of strain ONBA-17. The enzymatic activity responsible for ONBA removal was stable. About 56 ± 2.2% of the original activity remained when the extract was incubated at 30 °C for 144 h. Results of GC analysis show that no objective substance could be detected in the mixture containing intra-cellular fraction solution sampled at 2 h. Moreover, there was no downtrend of ONBA concentration in the mixture containing extra-cellular and membrane fraction solutions. These results indicate that enzyme(s) involved in the initial degradation of ONBA was endoenzyme(s).
As shown in Figure 3, 85.2 ± 1.5% of ONBA was degraded by extract with induction. Meanwhile, in the treatment of non-induced, 86.5 ± 1.6% was metabolized. In controls, abiotic degradation was negligible. These results clearly indicate that there was no significant difference in ONBA degradation between induced and non-induced cells, suggesting that enzyme(s) responsible for the degradation might be constitutively expressed.
The degradative enzyme(s) exceeded 65.2 ± 1.3% of its relative activity in a pH range between 6.4 and 8.0, with an optimum at pH 7.4. Enzyme(s) was stable in a pH range between 6.0 and 8.6. The enzyme(s) was optimally active at around 32 °C and fairly stable under 45 °C. These were logical, because the optimum pH value and growth temperature of the bacterium were around above conditions. It was nearly completely inactivated at 60 °C. Furthermore, the effects of various chemicals on enzymatic activity were listed in Table 1. No significant effect (less than 5% inhibition or activation) was observed in the presence of Fe2+, Ba2+, Cd2+ and Zn2+. The presence of 0.2 mM Li+, Ni2+, Co2+ and Fe3+ caused more than 20% enzyme activity inhibition. Addition of some divalent metal ions (Mg2+, Ca2+ and Mn2+) indeed enhanced the activity (> 15%). The presence of Tween 20 and Tween 80, and chelating agent EDTA resulted in a 46-63% inhibition of the activity. The strongest inhibition was observed with addition of surfactant SDS. The activity was fully depressed.
Metabolism by whole cells
During the degradation of ONBA several degradation products as well as some unknown transient accumulative metabolites were detected. Among these, existence of 2-nitrobenzoic acid and 2, 3-dihydroxybenzoic acid was confirmed. 2, 3-dihydroxybenzoic acid was firstly identified by characteristic mass fragments as follows: molecular ion [M+ (% relative intensity)] at m/z 370 (21) with major fragment ions at m/z 355 (45), 73 (100). Subsequently, this was further proved by comparison to the standard. Furthermore, ONBA-grown cells of strain P. putida ONBA-17 rapidly metabolized the substance with concomitant nitrite release. However, ammonia was not detected in the medium. Nitrite release was nearly stoichiometric when the cells used in the assay were grown in the presence of NH4C1 (data not shown). Extracts of cells grown on ONBA exhibited catechol 2, 3-dioxygenase activity but not catechol 1, 2-dioxygenase activity with catechol and 2, 3-dihydroxybenzoic acid (data not shown). Based on above results, we would like to propose the pathway of ONBA degradation by the bacterium as shown in Figure 4, although on their own they could not be served as conclusive evidences due to the possibilities of either multiple overlapping pathways being present in the bacterium or non-specificity of action of ONBA-induced enzymes as previously pointed out by Rhys-Williams et al. (1993). In addition, we have not been able to show growth of ONBA-17 on the postulated metabolites, 2-nitrobenzoic acid and 2, 3-dihydroxybenzoic acid. This might be due either to the toxicity or to the inability of the substances to act as inducers of the catabolic enzymes.
In short, this study indicates that ONBA-17 was a member of P. putida. Effects of inoculum amount, incubation condition and initial concentrations on ONBA biodegradation were investigated. The enzyme(s) involved was proved to be endoenzyme(s) and constitutively expressed. Besides, effects of different compounds on relative activity of degradative enzyme were examined. With GC-MS and enzyme assays, 2-nitrobenzoic acid and 2, 3-dihydroxybenzoic acid were detected as the metabolites of ONBA biodegradation, and relevant metabolic pathway was preliminary proposed. This is the first report, to our knowledge, on the deduction of ONBA degrading pathway within a bacterial strain affiliates to P. putida. The data obtained from this study could improve current understandings on nitro-substituted aromatics biodegradation. However, further research is still needed to clarify the whole course and key genes involved in the metabolism.
Acknowledgments
This work was supported by research center of Agricultural and Forestry Carbon Sinks and Ecological Environmental Remediation, China National Natural Science Foundation (grant no. 31100087), Zhejiang Provincial Natural Science Foundation (Cloning, expression and enzymatic properties analysis of 2-NBA metabolic genes, Y14C010026), and Qingnianbajian Program of Zhejiang Agricultural and Forestry University. We are grateful for their financial support.
Submitted: October 25, 2013
Approved: April 17, 2014
All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License CC BY-NC.
- Anwar S, Liaquat F, Khan QM, Khalid ZM, Iqbal S (2009) Biodegradation of chlorpyrifos and its hydrolysis product 3, 5, 6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J Hazard Mater 168:400-405.
- Chakraborty R, Wu CH, Hazen TC (2012) Systems biology approach to bioremediation. Curr Opin Biotechnol 23:483-490.
- Chen SH, Yang L, Hu MY, Liu JJ (2011) Biodegradation of fenvalerate and 3-phenoxybenzoic acid by a novel Stenotrophomonas sp. strain ZS-S-01 and its use in bioremediation of contaminated soils. Appl Microbiol Biotechnol 90:755-767.
- Grimont PAD (1999) Taxonomy and classification of bacteria. In: Manual of Clinical Microbiology, Murray, P.R., Baron, E.J., Pfaller, M.A., Tenover, F.C., Yolken, R.H. (eds). ASM Press, Washington, D.C., pp 249-259.
- Haigler BE, Spain JC (1993) Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl Environ Microbiol 59:2239-2243.
- Harold CN, Leon AH (1965) The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem 240:3685-3692.
- Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184-192.
- Hussain S, Arshad M, Saleem M, Khalid A (2007) Biodegradation of α- and β-endosulfan by soil bacteria. Biodegradation 18:731-740.
- Jeon CO, Madsen EL (2013) In situ microbial metabolism of aromatic-hydrocarbon environmental pollutants. Curr Opin Biotechnol 24:474-481.
- Lee KS, Parales JV, Friemann R, Parales RE (2005) Active site residues controlling substrate specificity in 2-nitrotoluene dioxygenase from Acidovorax sp. strain JS42. J Ind Microbiol Biotechnol 32:465-473.
- Liang WQ, Wang ZY, Li H, Wu PC, Hu JM, Luo N, Cao LX, Liu YH (2005) Purification and characterization of a novel pyrethroid hydrolase from Aspergillus niger ZD11. J Agr Food Chem 53:7415-7420.
- Liu C, Ali SW, Guan LB, Yu FB, Li SP, Wong MH (2012) Biotreatment of o-nitrobenzaldehyde manufacturing wastewater and changes in activated sludge flocs in a sequencing batch reactor. Bioresour Technol 104:228-234.
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with Folin-phenol reagent. J Biol Chem 193:265-275.
- Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109-118.
- Nishimori E, Kita-Tsukamoto K, Wakabayashi H (2000) Pseudomonas plecoglossicida sp. nov., the causative agent of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis Int J Syst Evol Microbiol 50:83-89.
- Perelo LW (2010) Review: in situ and bioremediation of organic pollutants in aquatic sediments. J Hazard Mater 177:81-89.
- Ramos JL, Marqués S, van Dillewijn P, Espinosa-Urgel M, Segura A, Duque E, Krell T, Ramos-González MI, Bursakov S, Roca A, Solano J, Fernádez M, Niqui JL, Pizarro-Tobias P, Wittich RM (2011) Laboratory research aimed at closing the gaps in microbial bioremediation. Trends Biotechnol 29:641-647.
- Rhys-Williams W, Taylor SC, Williams PA (1993) A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas J Gen Microbiol 139:1967-1972.
- Sambrook J, Fritsch EF, Maniatis T (2001) Molecular Cloning: a Laboratory Manual. 3rd edition. Cold Spring Harbor Laboratory, Cold Spring Harbor, pp 631-632.
- Verhille S, Baïda N, Dabboussi F, Hamze M, Izard D, Leclerc H (1999) Pseudomonas gessardii sp. nov. and Pseudomonas migulae sp. nov., two new species isolated from natural mineral waters. Int J Syst Bacteriol 49:1559-1572.
- Xu JH, Hong Q, Hong YF, Li SP (2011) Isolation, identification and degradation of carbofuran-degrading strain CFDS-1. China J Appl Environ Biol 17:237-242.
- Yu FB, Ali SW, Guan LB, Li SP, Zhou S (2010) Bioaugmentation of a sequencing batch reactor with Pseudomonas putida ONBA-17, and its impact on reactor bacterial communities. J Hazard Mater 176:20-26.
- Yu FB, Guan LB, Zhou S (2008) Isolation and preliminary characterization of a o-nitrobenzaldehyde-degrading Alcaligenes sp. ND1. Braz J Microbiol 39:776-779.
- Yu FB, Shan SD, Luo LP, Guan LB, Qin H (2013) Isolation and characterization of a Sphingomonas sp. strain F-7 degrading fenvalerate and its use in bioremediation of contaminated soil. J Environ Sci Heal B 48:198-207.
- Yu FB, Shen B, Li SP (2006) Isolation and characterization of Pseudomonas sp. strain ONBA-17 degrading o-nitriobenzaldehyde. Curr Microbiol 53:457-461.
Publication Dates
-
Publication in this collection
13 Feb 2015 -
Date of issue
Dec 2014
History
-
Received
25 Oct 2013 -
Accepted
17 Apr 2014