Abstract
Two Leptospira sp. isolates were obtained by the first time from goats in Brazil and characterized by sequencing rrs, rpoB and secY genes, PFGE and typing with monoclonal antibodies. Both isolates are identical and belong to Leptospira santarosai. Analysis of the rrs and the rpoB genes sequences revealed 100% identity between the goat isolates and the Bananal reference strain. When secY sequences of the two isolates were compared to each other, it was observed that they had identical sequences. However, when compared to that of the Bananal reference strain, there were 15 mismatches along the 549 bp secY sequence. In conclusion, molecular methods are increasingly useful for the characterization of leptospires and allowed to identify those isolates of caprine origin as closely related but not identical to serovar Bananal, and constitute a new type named Carioca.
Leptospira; serogrouping; sequencing; PFGE; rrs
RESEARCH PAPER
Molecular characterization of the first leptospires isolated from goats in Brazil
Walter LilenbaumI; Frederico KremerII; Paula RistowIII; Odir DellagostinII; Pascale BourhyIV; Rudy HartskeerlV; Silvio VasconcellosVI
ILaboratório de Bacteriologia Veterinária, Universidade Federal Fluminense, Niterói, RJ, Brazil
IINúcleo de Biotecnologia, Centro de Desenvolvimento Tecnológico, Universidade Federal de Pelotas, Pelotas, RS, Brazil
IIIInstituto de Biologia, Universidade Federal da Bahia, Salvador, BA, Brazil
IVInstitut Pasteur, Unité de Biologie des Spirochètes, Paris, France
VKIT Biomedical Research, Royal Tropical Institute, WHO/FAO/OIE and National Leptospirosis Reference Center, Amsterdam, Netherlands
VILaboratório de Zoonoses Bacterianas, Faculdade de Medicina Veterinária e Zootecnia, Universidade de São Paulo, São Paulo, Brazil
Correspondence Correspondence : W. Lilenbaum Laboratório de Bacteriologia Veterinária Rua Hernani Mello 101, sala 309 Universidade Federal Fluminense 24210-130 Rio de Janeiro, RJ, Brazil E-mail: mipwalt@vm.uff.br
ABSTRACT
Two Leptospira sp. isolates were obtained by the first time from goats in Brazil and characterized by sequencing rrs, rpoB and secY genes, PFGE and typing with monoclonal antibodies. Both isolates are identical and belong to Leptospira santarosai. Analysis of the rrs and the rpoB genes sequences revealed 100% identity between the goat isolates and the Bananal reference strain. When secY sequences of the two isolates were compared to each other, it was observed that they had identical sequences. However, when compared to that of the Bananal reference strain, there were 15 mismatches along the 549 bp secY sequence. In conclusion, molecular methods are increasingly useful for the characterization of leptospires and allowed to identify those isolates of caprine origin as closely related but not identical to serovar Bananal, and constitute a new type named Carioca.
Key words: Leptospira, serogrouping, sequencing, PFGE, rrs.
Introduction
Leptospirosis in goats constitutes one of the major infectious diseases, compromising reproductive efficiency, and, consequently, dairy and meat productivity in goat breeding (Martins et al., 2012). Besides that, leptospirosis is one of the most important zoonosis in the world and the real role of domestic species as reservoirs remains to be elucidated. Taxonomy of leptospires is still controversy, since both serological and molecular classification are accepted, but they very often do not match. Therefore, molecular classification of leptospiral isolates are increasingly important, and should be encouraged (Cerqueira and Picardeau, 2009). In 2007, members of our group (WL, SAV) reported the first isolation of leptospires from goats in Brazil (Lilenbaum et al., 2007). In that study two isolates of Leptospira sp. (identified as M72/06-6 and M72/06-13) were obtained from animals from different flocks. At that moment, only presumptive serological identification was conducted, suggesting that both strains belong to serogroup Grippotyphosa. The aim of this study is to describe the molecular characterization of those leptospiral isolates.
Material and Methods
Isolate M72/06-6 was recovered from an asymptomatic female dairy goat that was seroreactive against serogroup Grippotyphosa (Microscopic agglutination test - MAT titre of 400 against serovar Grippotyphosa). The owner reported occasional flooding during rainy season, what was assumed to be the most probable source of the infection. The other isolate (M72/06-13) was recovered from a male goat, with no clinical signs or leptospirosis and without detectable reproductive disorders. It presented a MAT titer of 200 against serovar Grippotyphosa.
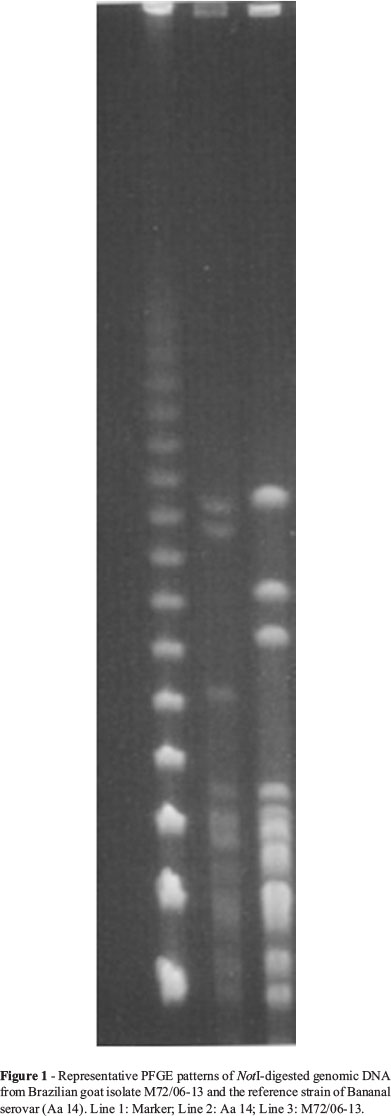
For pulsed-field gel electrophoresis (PFGE), cells were embedded in agarose plugs as previously described (Herrmann et al., 1992) and the DNA in the plugs digested with NotI. PFGE was performed in a contour-clamped homogeneous electric field DRII apparatus (Bio-Rad Laboratories, Richmond, CA). Restriction fragments were resolved with ramping from 5 to 60 s for 50 h, 1 to 30 s for 40 h, or from 1 to 70 s for 36 h at 6 V/cm.
Molecular characterization of the isolates was conducted by sequencing rrs, rpoB and secY genes. Briefly, leptospiral cultures were centrifuged at 12,000 rpm for 20 min at 4 °C and genomic DNA was extracted from cell pellets using the illustra bacteria genomicPrep mini spin kit (GE Healthcare), following the protocol for Gram negative bacteria. The rpoB gene was amplified by PCR using primers Lept 1900f and Lept 2500r, following the protocol described by La Scola et al. (La Scola et al., 2006). PCR amplification of rrs gene was carried out with universal primers (Weisburg et al., 1991) and sequencing was performed using internal primers previously described (Silva et al., 2007). The secY gene was amplified and sequenced as previously described (Ahmed et al., 2009). DNA sequencing was performed in a MegaBACE 1000 DNA sequencer (GE Healthcare), with Dynamic ET-terminator technology. The sequences obtained were submitted to BLAST alignment (www.ncbi.nlm.nih.gov/BLAST) against sequences available in GenBank.
Isolates were further typed at the serovar level by performing MAT with panels of the 13 monoclonal antibodies (mAbs) that characteristically agglutinate serovars belonging to the specific serogroup Grippotyphosa (F71C2, F71C3, F71C9, F71C13, F71C16, F71C17, F164C1, F165C1, F165C2, F165C3, F165C7, F165C8 and F165C12), as described (Feresu et al., 1995) .
PFGE analysis of NotI-digested genomic DNA revealed at least three distinct patterns for the typed isolates (Figure 1).
Results
Sequencing of the three genes (rrs, secY and rpoB) revealed that both strains belong to the pathogenic species Leptospira santarosai. Analysis of the rrs and the rpoB genes sequences revealed 100% identity between the goat isolates and the Bananal reference strain. Sequencing of secY gene from isolates M72/06-6 and M72/06-13, and for the Bananal reference strain were obtained and deposited in GenBank under accession numbers KF366253, KF311106 and KF366254, respectively. When secY sequences of M72/06-6 and M72/06-13 were compared to each other, it was observed that both isolates had identical sequences. However, when compared to that of the Bananal reference strain, there were 15 mismatches along the 549 bp secY sequence (Figure 2).
Agglutination titers of all mAbs classified the two isolates as members of serogroup Grippotyphosa and suggested that both isolates are serologically identical and much similar, but not identical to serovar Bananal. Titers were detected for F71C2 (10240), F71C9 (5120), F71C17 (5120), F165C1 (20480) and F165C3 (20480).
Discussion
Results from serogrouping were most in concordance with serovar Bananal of serogroup Grippotyphosa. This serovar was first isolated in the region named Bananal, distant 275 km from the studied region (Cordeiro et al., 1981). Therefore, considering the serological and molecular results obtained, as well as the epidemiological data, it was assumed that both strains were closely related to serovar Bananal. There was no evidence of exchange of animals between the two flocks. Since the two farms were located in a distance of about 15 km, it is possible to infer that animals from the different flocks had a similar exposure, probably environmental.
It is noteworthy that although serological methods were able to determine the serogroup of the isolates, it was impossible to unambiguously distinguish between the Bananal reference strain and the two isolates only employing serogrouping. PFGE has been the gold standard method for genotyping Leptospira strains (Bourhy et al., 2013). In the present study, molecular tools, mainly PFGE and particularly secY sequencing, demonstrated important genetic polymorphisms between the isolates and Bananal reference strain. Clearly serogrouping alone cannot clearly differentiate between closely related isolates, and the classification of leptospires benefits from the use of molecular tools, as was shown in other studies (Ahmed et al., 2012). Nonetheless, there is an urgent need for a definitive method for leptospiral typing. One limitation of molecular methods is that, in contrary of serological methods, as the complex, laborious and expensive cross agglutination absorption test (CAAT), genetic studies does not allow us to classify those strains as a new serovar, but only as a genetically diverse strain, or genotype. Nevertheless, the association of agglutination tests with molecular analysis (PFGE and secY sequencing) allowed us to discriminate between the isolates and the reference strains. We suggest naming these genetically diverse strains as Carioca.
Conclusion
In conclusion, molecular methods are increasingly useful for the characterization of leptospires and allowed to identify the first isolates in Brazil of caprine origin as closely related but not identical to serovar Bananal, and constitute a new type named Carioca.
Acknowledgments
This study was supported by FAPERJ. We thank Dr. M. Picardeau, A. Landier, S. Bremont (Institut Pasteur), F.K. Seixas (UFPel) and F. Z. Brandão (UFF) for the technical assistance.
Submitted: November 01, 2013
Approved: April 17, 2014
All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License CC BY-NC.
- Ahmed A, Engelberts MF, Boer KR, Ahmed N, Hartskeerl RA (2009) Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLoS ONE 4:e7093.
- Ahmed A, Grobush MP, Klatser P, Hartskeerl RA (2012) Molecular approaches in the detection and characterization of Leptospira J Bacteriol Parasitol 3:133.
- Bourhy P, Herrmann Storck C, Theodose R, Olive C, Nicolas M, Hochedez P, Lamaury I, Zinini F, Brémont S, Landier A, Cassadou S, Rosine J, Picardeau M (2013) Serovar diversity of pathogenic Leptospira circulating in the French West Indies. PLoS Negl Trop Dis 7:e2114.
- Cerqueira GM, Picardeau M (2009) A century of Leptospira strain typing. Infect Genet Evol 9:760-768.
- Cordeiro F, Sulzer CR, Ramos AA (1981) Leptospira interrogans in several wildlife species in southeast Brazil. Pesq Vet Bras 1:19-29.
- Feresu SB, Bolin CA, Korver H, Van der Kemp H (1995) Identification of leptospires of the Pomona and Grippotyphosa serogroups isolated from cattle in Zimbabwe. Res Vet Sci 89:92-94.
- Herrmann JL, Bellenger E, Perolat P, Baranton G, Saint Girons I (1992) Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J Clin Microbiol 30:1696-1702.
- La Scola B, Bui LT, Baranton G, Khamis A, Raoult D (2006) Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263:142-147.
- Lilenbaum W, Morais ZM, Gonçales AP, Souza GO, Richtzenhain L, Vasconcellos SA (2007) First isolation of leptospires from dairy goats in Brazil. Braz J Microbiol 38:507-510.
- Martins G, Penna B, Hamond C, Leite RC, Silva A, Ferreira A, Brandão F, Oliveira F, Lilenbaum W (2012) Leptospirosis as the most frequent infectious disease impairing productivity in small ruminants in Rio de Janeiro, Brazil. Trop Anim Health Prod 44:773-777.
- Silva EF, Brod CS, Cerqueira GM, Bourscheidt D, Seyffert N, Queiroz A, Santos CS, Ko AI, Dellagostin AO (2007) Isolation of Leptospira noguchii from sheep. Vet Microbiol 121:144-149.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697-703.
Correspondence
Publication Dates
-
Publication in this collection
13 Feb 2015 -
Date of issue
Dec 2014
History
-
Received
01 Nov 2013 -
Accepted
17 Apr 2014