Abstract
A modified colorimetric high-throughput screen based on pH changes combined with an amidase inhibitor capable of distinguishing between nitrilases and nitrile hydratases. This enzymatic screening is based on a binary response and is suitable for the first step of hierarchical screening projects.
nitrile hydratase; amidase; nitrilase; amidase inhibitor; high-throughput screening
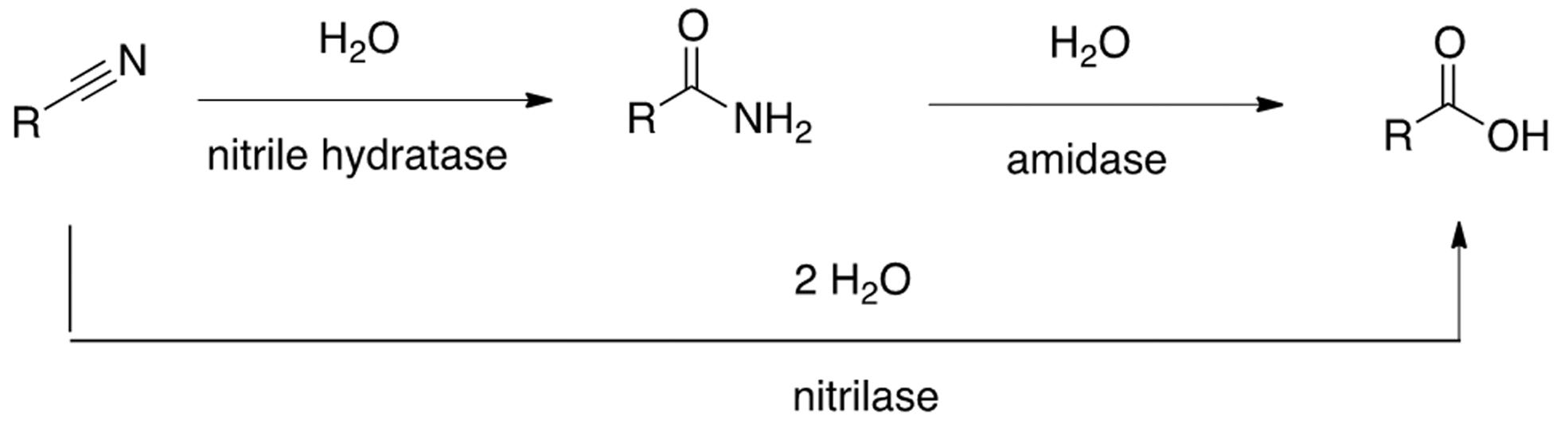
Nitrile metabolizing enzymes are of outstanding synthetic importance because of their ability to afford amides, carboxylic acids, amines and cyanohydrins, compounds used as building blocks in the pharma and chemical industries (van Pelt 2011van Pelt S, Zhang M, Otten LG, Holt DY, Sorokin F, van Rantwijk, Black GW, Perry JJ, Sheldon RA (2011) Probing the enantioselectivity of a diverse group of purified cobalt-centred nitrile hydratases. Org. Biomol. Chem 9:3011–3019., Chen 2009Chen J, Zheng RC, Zheng YG, Shen YC (2009) Microbial transformation of nitrile to high-value acids and amides. Adv. Biochem. Engin/Biotechnol 113: 33–77., Lin 2012Lin Z-J, Zheng R-C, Wang Y-J, Zheng Y-G, Shen Y-C (2012) Enzymatic production of 2-amino-2,3-dimethylbutyramide by cyanide-resistant nitrile hydratase. J Ind Microbiol Biotechnol 39:133–141., Yamada 2001Yamada H, Shimizu S, Kobayashi M (2001) Hydratases involved in nitrile conversion: screening, characterization and application. The Japan Chemical Journal Forum and John Wiley & Sons Inc. The Chemical Record 1:152–161.). They can also be potent biocatalysts for detoxification of anthropogenic toxic nitrile pollutants that are widespread throughout the world (Vesela 2012Vesela AB, Pelantova H, Sulc M, Mackova M, Lovecka P, Thimova M, Pasquarelli F, Picmanova M, Patek M, Bhalla TC, Martinkova L (2012) Biotransformation of benzonitrile herbicides via the nitrile hydratase-amidase pathway in rhodococci. J Ind Microbiol Biotechnol 39:1811–1819., Vesela 2010Vesela, AB, Franc M, Pelantová H, Kubac D, Vejvoda V, Sulc M, Bhalla TC, Mackova M, Lovecka P, Janu P, Demmerova K, Martinkova L (2010) Hydrolysis of benzonitrile herbicides by soil actinobacteria and metabolite toxicity. Biodegradation 21:761–770.). Among them, nitrile hydratase is an interesting commercial enzyme already used successfully on a ton scale for the production of commodities and pharmaceuticals such as acrylamide and nicotinamide (Prasad 2010Prasad S, Bhalla TC (2010) Nitrile hydratases (NHases): At the interface of academia and industry. Biotech Adv 28:725–741., Kobayashi 2000Kobayashi M, Shimizu S (2000) Nitrile hydrolases. Curr Opin Chem Biol 4:95–102., Yamada 2001Yamada H, Shimizu S, Kobayashi M (2001) Hydratases involved in nitrile conversion: screening, characterization and application. The Japan Chemical Journal Forum and John Wiley & Sons Inc. The Chemical Record 1:152–161.). Nitrile hydratase was first discovered in 1980 by Yasano and collaborators and it is believed that for the most part, they are involved in a cascade reaction with amidases, affording carboxylic acids from nitriles passing through an amide intermediate (Yasano 1980Yasano Y, Tani T, Yamada H (1980) A new enzyme “nitrile hydratase” which degrades acetonitrile in combination with amidase. Agric Biol Chem 44:2251–2252.). Nitrilases are also present in many different species and afford a carboxylic acid directly from a nitrile compound (Prasad 2010Prasad S, Bhalla TC (2010) Nitrile hydratases (NHases): At the interface of academia and industry. Biotech Adv 28:725–741.) (Figure 1).
A number of screening assays for nitrile-converting enzymes based on continuous and stopped methods are well documented in the literature (Asano 2002Asano Y (2002) Overview of screening for new microbial catalysts and their uses in organic synthesis – selection and optimization of biocatalysts J Biotechnol 94:65–72., Martinkova 2008Martinkova L, Vejvoda V, Kren V (2008) Selection and screening for enzymes of nitrile metabolism. J Biotechnol 133: 318–326.; Reisinger 2006Reisinger C, Assema F, Schürmann M, Hussain Z, Remler P, Schwab H (2006) A versatile colony assay based on NADH fluorescence. J Mol Catal B: Enzym 39:149–155., Santoshkumar 2010Santoshkumar M, Nayak AS, Anjaneya O, Karegoudar TB (2010) A plate method for screening of bacteria capable of degrading aliphatic nitrile. J Ind Microbiol Biotechnol 37:111–115.; He 2011He Y-C, Ma C-L, Xu J-H, Zhou L (2011) A high-throughput screening strategy for nitrile-hydrolyzing enzymes based on ferric hydroxamate spectrophotometry. App Microbiol Biotechnol 89:817–823.; Zheng 2011, Yazbeck 2006Yazbeck DR, Durao PJ, Xie Z, Tao J (2006) A metal ion-based method for the screening of nitrilases. J Mol Catal B: Enzym 39:156–159., Wang 2012Wang Y-J, Liu Z-Q, Zheng R-C, Xue Y-P, Zheng Y-G (2012) Screening, cultivation, and biocatalytic performance of Rhodococcus boritolerans FW815 with strong 2,2-dimethylcyclopropanecarbonitrile hydratase activity. J Ind Microbiol Biotechnol 39:409–417.). However, as nitrilases and nitrile hydratase-amidases afford the same final product, it is important to design a screening assay able to distinguish between the two enzymatic pathways.
Herein, we describe a colorimetric high-throughput screening assay based on pH changes coupled with the use of an amidase inhibitor. This screen is based on a binary response allowing differentiation between nitrilases and nitrile hydratase-amidases enzymatic systems and is suitable for the first step of hierarchical screening projects.
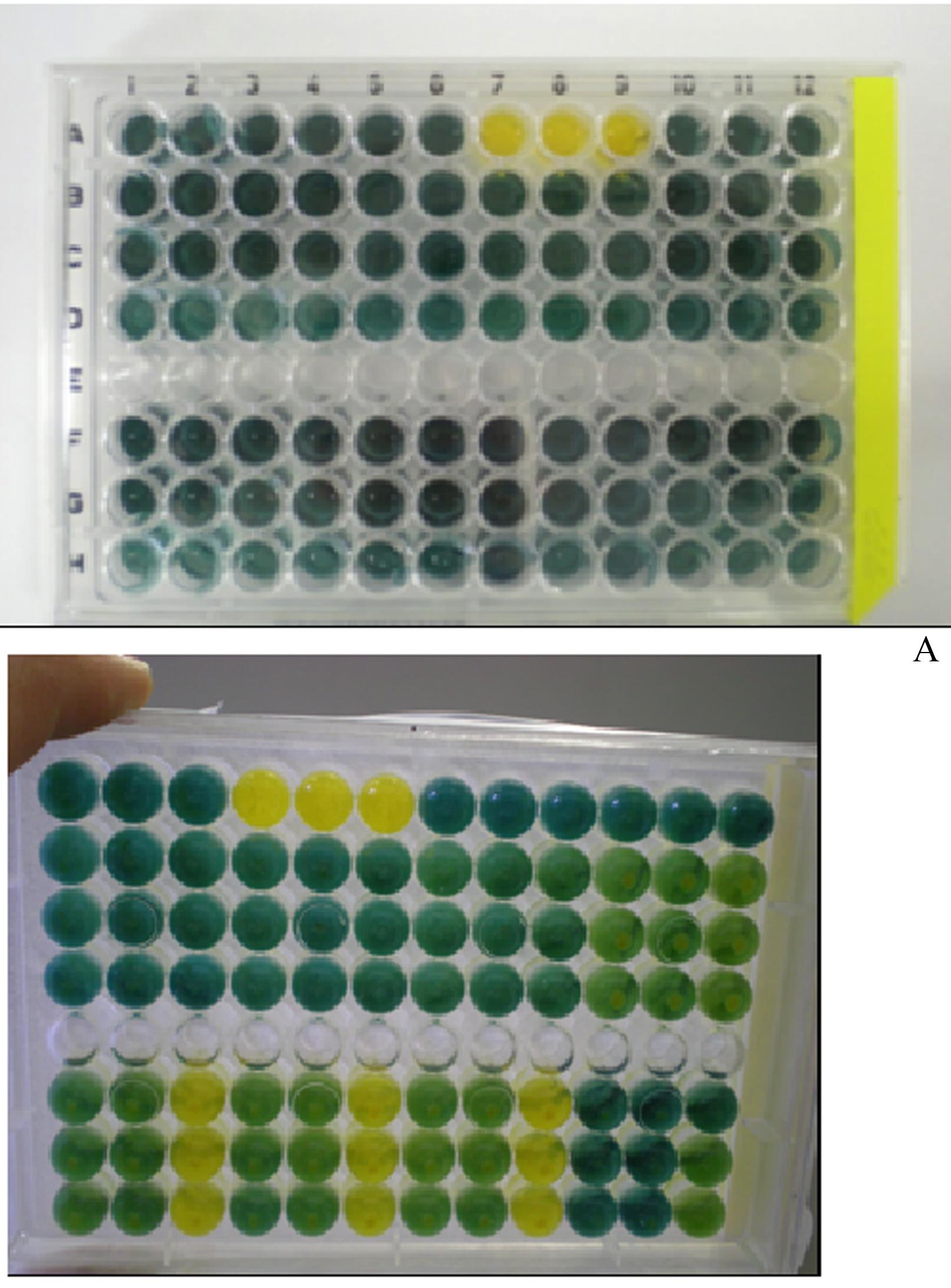
A Banerjee-modified colorimetric and pH sensitive assay coupled with an amidase inhibitor was performed for screening nitrilase and nitrile hydratase-amidase enzymes. Commercially available microorganisms potentially containing the nitrile hydratase and nitrilase enzymatic systems were used as positive controls. All nitriles and their corresponding amides and carboxylic acids and an amidase inhibitor were evaluated to detect any possible color change interferences within the enzymatic assay system. It was assumed that the strains that did not accumulate the amide during nitrile degradation indicated nitrilase activity. The intermediate accumulation of the corresponding amide during nitrile metabolism combined with carboxylic acid formation was taken as an indication of the existence of a nitrile hydratase-amidase system (Layh 1997Lay N, Hirrlinger B, Stolz A, Knackmuss HJ (1997) Enrichment strategies for nitrile-hydrolysing bacteria. App Microbiol Biotechnol 47:668–674.). The expression of nitrile hydratases was induced by acetonitrile or benzonitrile for aliphatic and aromatic nitriles, respectively. Mandelonitrile was too toxic to the microorganisms prior to enzymatic induction therefore it was not used as an inducing agent. The well-known amidase inhibitor diethyl phosphoramidate, DEPA (Bauer 1998Bauer R, Knackmuss HJ, Soltz A (1998) Enantioselective hydration of 2-arylpropionitriles by a nitrile hydratase from Agrobacterium tumefasciens strain d3. Appl Microbiol Biotechnol 49:89–95.), was chosen for this screening since its color did not affect the assay readout and also it is not influenced by pH changes during the course of the assay. The use of an amidase inhibitor permitted the accumulation of the amide intermediate, thus allowing the discrimination between nitrile hydratase-amidases and nitrilases when only one of these enzymatic systems was present (Brady 2004Brady, D, Beeton A, Zeevaart J, Kgaje C, van Rantwijk F, Sheldon RA (2004) Characterization of nitrilase and nitrile hydratase biocatalytic systems. Appl Microbiol Biotechnol 64:76–85.). However, when the microorganism had both enzymatic systems, it was not possible to reach a definitive conclusion. Moreover, a microbial control experiment is important since the production and/or excretion of acidic metabolites into the extracellular media in concentrations high enough to cause color changes in the pH indicator may compromise the assay validity. The screening assays could be monitored by simple microtiter plate visual inspection (Figure 2). Additionally, a colorimetric master plate was used as reference color scale.
Screening for nitrile hydratase and nitrilase producing strains using mandelonitrile as a substrate in a microplate. Row A: control experiments: A1–A3 mandelonitrile, A4–A6: mandelamide, A7–A9: mandelic acid; A10–A12: DEPA. A: time zero; B: time = 36 h. * Photo was taken from the bottom up. Rows B, C, D, F, G and H: different isolated microorganisms were checked for their ability to hydrolyze mandelonitrile
As expected, Pseudomonas putida CCT 2357 and Pseudomonas fluorescens CCT 3178 are exclusively nitrilase producing strains (Table 1). This result is supported by evidence from the literature (Chen 2009Chen J, Zheng RC, Zheng YG, Shen YC (2009) Microbial transformation of nitrile to high-value acids and amides. Adv. Biochem. Engin/Biotechnol 113: 33–77., Prasad 2010Prasad S, Bhalla TC (2010) Nitrile hydratases (NHases): At the interface of academia and industry. Biotech Adv 28:725–741.) and can be rationalized by the maintenance of yellow color in the presence or absence of amidase inhibitor. On the other hand, Nocardia simplex CCT 3022 produces only nitrile hydratase-amidase enzymes since the carboxylic acid formation was detected in the experiment without amidase inhibitor but not in the assay with the addition of DEPA. If a nitrilase was present, color change would be expected in the experiment with DEPA addition, however, no color change was observed. The strains Rhodococcus ruber CCT 1879, Rhodococcus equi CCT 0541, Rhodococcus erythropolis CCT 1878 and Nocardia brasiliensis CCT 3439 produce both a nitrilase and nitrile hydratase-amidase. In this case, the screening assay cannot give a conclusive response. Experiments were completed in the presence of a nitrilase inhibitor (AgNO3), however, they were unsuccessful because of the high sensitivity of AgNO3 to air and the formation of colored precipitate. The results obtained with Arthrobacter sp CCT 1875 show the importance of microbial control. This screening assay is not suited to cases where acidic metabolites are produced or excreted into extracellular media in concentrations high enough to cause a pH change above the buffer capacity.
Enzymatic assay results for detection of nitrilase, nitrile hydratase and amidase activities.
The reactions were performed on a 10 mL scale in an Erlenmeyer flask and monitored by GC-FID to confirm the results obtained with the microtiter plate assay (Figures S1 Figure S1 Chromatogram of acetonitrile (rt 1.90 min), acetamide (rt 9.38 min) and acetic acid (rt 5.60 min). Temperature program: 35 °C (4 min) 25 °C/min to 280 °C (3 min). Split ratio 1: 100. and S2: Supporting Information Figure S2 Chromatogram of mandelonitrile (rt 7.45 min), mandelamide (rt 8.60 min) and methyl mandelate (rt 7.21 min). Temperature program: 80 °C (3 min) 25 °C/min to 300 °C (3 min). Split ratio 1: 100. ). No false positives or false negatives were detected.
To confirm that this method can be used for screening of nitrile hydratase-producing microorganisms, we used it to screen our in house wild type microorganism library. These microorganisms were previously isolated from different sources (cassava waste water, cassava, benzonitrile herbicide impacted soils and UNESP garden) by enrichment techniques. From a library of 130 microorganisms isolated from nitrile-impacted and non-impacted areas, 19 positive hits were detected, of which 12 were nitrilase producers, 3 were NHase-amidase producers and 5 produced both enzymatic systems.
Interestingly, all microbial cultures which afforded positive hits came from nitrile-impacted areas. These results suggest the importance of natural microbial acclimation and also suggest a strategy for enzyme identification. The enzymatic assay was capable of detecting and distinguishing nitrile hydratases from nitrilases when only one was produced in a bacterial strain. The enzymatic assay was validated by GC-FID monitoring. Nitrile maximum inhibitory concentrations (MIC) were determined using a MTT assay. The MIC values varied for each tested nitrile and microbial culture. The wild type microorganisms are currently being subjected to 16S RNA identification and show promise as whole cell biocatalysts.
Acknowledgement
The authors thank Sao Paulo Research Foundation FAPESP for financial support.
References
- Asano Y (2002) Overview of screening for new microbial catalysts and their uses in organic synthesis – selection and optimization of biocatalysts J Biotechnol 94:65–72.
- Banerjee AB, Kaul P, Sharma R, Banerjee UC (2003) A high-throughput amenable colorimetric assay for enantioselective screening of nitrilase-producing microorganisms using pH sensitive indicators. J Biomol Screen 8:559–565.
- Bauer R, Knackmuss HJ, Soltz A (1998) Enantioselective hydration of 2-arylpropionitriles by a nitrile hydratase from Agrobacterium tumefasciens strain d3. Appl Microbiol Biotechnol 49:89–95.
- Brady, D, Beeton A, Zeevaart J, Kgaje C, van Rantwijk F, Sheldon RA (2004) Characterization of nitrilase and nitrile hydratase biocatalytic systems. Appl Microbiol Biotechnol 64:76–85.
- Chen J, Zheng RC, Zheng YG, Shen YC (2009) Microbial transformation of nitrile to high-value acids and amides. Adv. Biochem. Engin/Biotechnol 113: 33–77.
- He Y-C, Ma C-L, Xu J-H, Zhou L (2011) A high-throughput screening strategy for nitrile-hydrolyzing enzymes based on ferric hydroxamate spectrophotometry. App Microbiol Biotechnol 89:817–823.
- Kobayashi M, Shimizu S (2000) Nitrile hydrolases. Curr Opin Chem Biol 4:95–102.
- Lay N, Hirrlinger B, Stolz A, Knackmuss HJ (1997) Enrichment strategies for nitrile-hydrolysing bacteria. App Microbiol Biotechnol 47:668–674.
- Lin Z-J, Zheng R-C, Wang Y-J, Zheng Y-G, Shen Y-C (2012) Enzymatic production of 2-amino-2,3-dimethylbutyramide by cyanide-resistant nitrile hydratase. J Ind Microbiol Biotechnol 39:133–141.
- Lin Z-J, Zheng RC, Lei L-H, Zheng Y-G, Shen Y-C (2011) Ferrous and ferric ions-based high-throughput screening strategy for nitrile hydratase and amidase J Microbiol Meth 85:214–220.
- Martinkova L, Vejvoda V, Kren V (2008) Selection and screening for enzymes of nitrile metabolism. J Biotechnol 133: 318–326.
- Prasad S, Bhalla TC (2010) Nitrile hydratases (NHases): At the interface of academia and industry. Biotech Adv 28:725–741.
- Reisinger C, Assema F, Schürmann M, Hussain Z, Remler P, Schwab H (2006) A versatile colony assay based on NADH fluorescence. J Mol Catal B: Enzym 39:149–155.
- Santoshkumar M, Nayak AS, Anjaneya O, Karegoudar TB (2010) A plate method for screening of bacteria capable of degrading aliphatic nitrile. J Ind Microbiol Biotechnol 37:111–115.
- van Pelt S, Zhang M, Otten LG, Holt DY, Sorokin F, van Rantwijk, Black GW, Perry JJ, Sheldon RA (2011) Probing the enantioselectivity of a diverse group of purified cobalt-centred nitrile hydratases. Org. Biomol. Chem 9:3011–3019.
- Vesela AB, Pelantova H, Sulc M, Mackova M, Lovecka P, Thimova M, Pasquarelli F, Picmanova M, Patek M, Bhalla TC, Martinkova L (2012) Biotransformation of benzonitrile herbicides via the nitrile hydratase-amidase pathway in rhodococci. J Ind Microbiol Biotechnol 39:1811–1819.
- Vesela, AB, Franc M, Pelantová H, Kubac D, Vejvoda V, Sulc M, Bhalla TC, Mackova M, Lovecka P, Janu P, Demmerova K, Martinkova L (2010) Hydrolysis of benzonitrile herbicides by soil actinobacteria and metabolite toxicity. Biodegradation 21:761–770.
- Wang Y-J, Liu Z-Q, Zheng R-C, Xue Y-P, Zheng Y-G (2012) Screening, cultivation, and biocatalytic performance of Rhodococcus boritolerans FW815 with strong 2,2-dimethylcyclopropanecarbonitrile hydratase activity. J Ind Microbiol Biotechnol 39:409–417.
- Yamada H, Shimizu S, Kobayashi M (2001) Hydratases involved in nitrile conversion: screening, characterization and application. The Japan Chemical Journal Forum and John Wiley & Sons Inc. The Chemical Record 1:152–161.
- Yasano Y, Tani T, Yamada H (1980) A new enzyme “nitrile hydratase” which degrades acetonitrile in combination with amidase. Agric Biol Chem 44:2251–2252.
- Yazbeck DR, Durao PJ, Xie Z, Tao J (2006) A metal ion-based method for the screening of nitrilases. J Mol Catal B: Enzym 39:156–159.
Supplementaty Material
Figure S1 Chromatogram of acetonitrile (rt 1.90 min), acetamide (rt 9.38 min) and acetic acid (rt 5.60 min). Temperature program: 35 °C (4 min) 25 °C/min to 280 °C (3 min). Split ratio 1: 100. Figure S2 Chromatogram of mandelonitrile (rt 7.45 min), mandelamide (rt 8.60 min) and methyl mandelate (rt 7.21 min). Temperature program: 80 °C (3 min) 25 °C/min to 300 °C (3 min). Split ratio 1: 100.Publication Dates
-
Publication in this collection
31 Mar 2015 -
Date of issue
Jan-Mar 2015
History
-
Received
06 Aug 2013 -
Accepted
06 June 2014