Abstract
An indigenous bacterial strain capable of utilizing 2,4-dichlorophenoxyacetic acid as the sole carbon and energy source was isolated from a soil used for grown wheat with a long-term history of herbicide use in Beijing, China. The strain BJ71 was identified as Cupriavidus campinensis based on its 16S rRNA sequence analysis and morphological, physiological, and biochemical characteristics. The degradation characteristics of strain BJ71 were evaluated. The optimal conditions for 2,4-D degradation were as follows: pH 7.0, 30 °C, 3% (v/v) inoculum size, and an initial 2,4-D concentration of 350 mg L−1. Up to 99.57% of the 2,4-D was degraded under optimal conditions after 6 days of incubation. Strain BJ71 was also able to degrade quizalofop and fluroxypyr. This is the first report of a 2,4-D-degrader containing tfdA gene that can utilize these two herbicides. In a biodegradation experiment, 87.13% and 42.53% of 2,4-D (initial concentration, 350 mg kg−1) was degraded in non-sterile and sterilized soil inoculated with BJ71, respectively, after 14 days. The 2,4-D degradation was more rapid in a soil microcosm including BJ71 than in a soil microcosm without BJ71. These results indicate that strain BJ71 is a potential candidate for the bioremediation of soil contaminated with the herbicide 2,4-D.
2,4-D-degrading bacterial strain; bioremediation; Cupriavidus campinensis BJ71; degradation characteristics
Introduction
The post-emergence systemic herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) has been widely used to control dicotyledonous weeds in cereal and grass crops for more than 60 years. Although this herbicide has a short half-life in soil and aquatic environments, its extensive use is a cause for concern because of the potential threat to the environment and human health (Chinalia et al., 2007Chinalia FA, Regali-Seleghin MH, Correa EM (2007) 2,4-D toxicity: cause, effect and control. Terrestrial and Aquatic Environmental Toxicology 1:24–33.). The WHO (World Health Organization) has classified 2,4-D as a hormonal herbicide with level II toxicity. It is considered to be a carcinogenic agent affecting the liver and heart. It also affects the central nervous system, leading to convulsions (Garc'a et al., 2006Garc'a GB, Konjuh C, Duffard RO et al. (2006) Dopamine-β-hydroxylase immunohistochemical study in the locus coeruleus of neonate rats exposed to 2,4-dichlorophenoxyacetic acid through mother's milk. Drug Chem Toxicol 29:435–442.; Maloney and Waxman, 1999Maloney EK, Waxman DJ (1999) Trans-activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol Appl Pharmacol 161:209–218.). Because of the relatively high water solubility and low soil-absorption coefficient of the free acid of 2,4-D, it often contaminates the environment when it enters streams, rivers, or lakes after drainage of agricultural lands (Chinalia and Killham, 2006Chinalia FA, Killham KS (2006) 2,4-Dichlorophenoxyacetic acid (2,4-D) biodegradation in river sediments of northeast-Scotland and its effect on the microbial communities (PLFA and DGGE). Chemosphere 64:1675–1683.; El-Bestawy and Hans-Jorgen, 2007El-Bestawy E, Hans-Jorgen A (2007) Effect of nutrient amendments and sterilization on mineralization and/or biodegradation of 14C-labeled MCPP by soil bacteria under aerobic conditions. Int Biodeterior Biodegradation 59:193–201.; Lane, 1991Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt, E., Goodfellow, M. (Eds.). Nucleic Acid Techniques in Bacterial Systematics. Wiley, New York, pp.115–175.). Most of the 2,4-D-degrading bacteria characterized to date are members of genera belonging to the β and γ subdivisions of the class Proteobacteria, and have been isolated from 2,4-D-treated environments (Gonzalez et al., 2012Gonzalez AJ, Gallego A, Gemini VL et al. (2012) Degradation and detoxification of the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) by an indigenous Delftia sp. strain in batch and continuous systems. Int Biodeterior Biodegradation 66:8–13.; Kamagata et al., 1997Kamagata Y, Fulthorpe RR, Tamura K et al. (1997) Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol 63:2266–2272.; Lee et al., 2005Lee T, Kurata S, Nakatsu C et al. (2005) Molecular analysis of bacterial community based on 16S rDNA and functional genes in activated sludge enriched with 2,4-dichlorophenoxyacetic acid (2,4-D) under different cultural conditions. Microb Ecol 49:151–162.). The best-described pathway for 2,4-D degradation is the plasmid-encoded pathway of the bacterium Ralstonia eutropha (formerly Alcaligenes eutrophus, Waustersia eutropha) strain JMP134 (Fukumori and Hausinger, 1993aFukumori F, Hausinger RP (1993a) Alcaligenes eutrophus JMP134 "2,4-dichlorophenoxyacetate monooxygenase" is an α-ketoglutarate-dependent dioxygenase. J Bacteriol 175:2083–2086.; Fukumori and Hausinger, 1993bFukumori F, Hausinger RP (1993b) Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J Biol Chem 268:24311–24317.; Streber et al., 1987Streber WR, Timmis KN, Zenk MH (1987) Analysis, cloning, and high-level expressions of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol 169:2950–2955.). Despite the many attempts to increase the biodegradation of 2,4-D by bioaugmentation of soils with degrading strains, no significant increases in pollutant removal compared with that in uninoculated soil have been achieved (Sánchez et al., 1994). The introduced strains do not always survive well in the soil environment because of various stresses, including competition with indigenous microorganisms (Manzano et al., 2007Manzano M, Morán AC, Tesser B et al. (2007) Role of eukaryotic microbiota in soil survival and catabolic performance of the 2,4-D herbicide degrading bacteria Cupriavidus necator JMP134. Antonie Van Leeuwenhoek 91:115–126.; Marrón-Montiel et al., 2006Marrón-Montiel E, Ordaz-Ruiz N, Rubio-Granados C et al. (2006) 2,4-D-degrading bacterial comsortium isolation kinetic characterization in batch and continuous culture and application for bioaugmenting an activated sludge microbial community. Process Biochem 41:1521–1528.). For successful bioremediation of herbicide-contaminated soil, it is necessary to construct a unique niche for the desired microbes so that they can be productively exploited (Singh, 2008Singh DK (2008) Biodegradation and bioremediation of pesticide in soil: concept, method and recent developments. Indian J Microbiol 48:35–40.). Hence, the use of native 2,4-D degrading microorganisms is a feasible strategy for the bioremediation of polluted sites. The purpose of this study was to isolate an indigenous 2,4-D degrading strain using enrichment techniques, and to evaluate the degradation characteristics of the strain under different conditions. Our long-term goal is to isolate and characterize an indigenous 2,4-D degrading strain for bioaugmentation during in situ clean-up of sites polluted with 2,4-D in China.
Materials and Methods
Chemicals
2,4-D, 2-methyl-4-chloro-phenoxyacetic acid (MCPA), mecoprop (99% purity), and eosin B were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ringers solution was purchased from Oxoid (Basingstoke, UK). Quizalofop, fluroxypyr, and all other chemicals were of analytical grade or higher purity.
Enrichment and isolation of 2,4-D-degrading bacteria
Soils used for enrichment of 2,4-D-degrading bacteria were collected from wheat fields in Beijing (40°25′40″ N, 116°82′79″ E), Henan Province (34°20′34″ N, 114°15′07″ E), and Qinghai Province (36°92′15″ N, 101°67′20″ E), China. These soils have been exposed to 2,4-D for at least 10 years. Ten grams of each soil was inoculated into an Erlenmeyer flask (250 mL) containing 100 mL minimal salt medium (MSM; containing MgSO4 0.2 g L−1, (NH4)2SO4 0.5 g L−1, KH2PO4 0.5 g L−1, K2HPO4 1.5 g L−1, Na2EDTA 0.12 g L−1, NaOH 0.02 g L−1, ZnSO4 0.004 g L−1, CuSO4 0.001 g L−1, Na2SO4 0.0001 g L−1, Na2MoO4 0.001 g L−1, CoCl2 0.0001 g L−1, MnSO4 0.0004 g L−1, and 0.5 mL concentrated H2SO4, pH 7.0) (Smejkal et al., 2001aSmejkal CW, Vallaeys T, Seymour FA et al. (2001a) Characterization of (R/S)-mecoprop [2-(2-methyl-4-chlorophenoxy)propionic acid]-degrading Alcaligenes sp. CS1 and Ralstonia sp. CS2 isolated from agricultural soils. Environ Microbiol 3:288–293.), which was supplemented with 500 mg L−1 2,4-D as the sole carbon and energy source. The flasks were incubated at 30 °C for 7 days in a rotary shaker at 150 rpm. Then, 10 mL enrichment culture showing degradation of 2,4-D was transferred to 100 mL fresh MSM containing 500 mg L−1 2,4-D and further incubated for 7 days. Four rounds of enrichment were performed and the final enrichment cultures were serially diluted and spread on MSM plates containing 500 mg L−1 2,4-D. After incubation at 30 °C for 4 days, single colonies with different morphologies were selected for further analysis of their degradation abilities. For each strain, the extent of degradation was determined by quantifying the amount of 2,4-D remaining in the culture by high-performance liquid chromatography (HPLC). One strain isolated from Beijing, designated as BJ71, degraded 2,4-D very quickly and showed the highest degradation rate of 2,4-D among the isolated strains. This strain was selected for further investigation. Isolates were stored frozen in 15% glycerol at −80 °C until analysis.
Identification and characterization of strain BJ71
Strain BJ71 was identified based on its 16S rRNA gene sequence analysis and morphological, physiological, and biochemical tests. Morphological, physiological, and biochemical tests were conducted according to Bergey's Manual of Systematic Bacteriology (Garrity et al., 2004Garrity GM, Bell JA, Lilburn TG (2004) Taxonomic outline of the prokaryotes Bergey's Manual of Systematic Bacteriology (second edition) Springer New York Berlin Heidelberg. doi: 10.1007/bergeysoutline 200405.
https://doi.org/10.1007/bergeysoutline20...
) and related documents (Vandamme and Coenye, 2004Vandamme P, Coenye T (2004) Taxonomy of the genus Cupriavidus: a tale of lost and found. Int J Syst Evol Microbiol 54:2285–2289.; Vaneechoutte et al., 2004Vaneechoutte M, K?mpfer P, De Baere T et al. (2004) Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al., 1990) comb. nov. Int J Syst Evol Microbiol 54:317–327.). Microscopic observation was performed with an HITACHI S3400N scanning electron microscope (Hitachi, Tokyo, Japan) at 20 kV. Samples were prepared for scanning electron microscopy by fixing bacterial colonies in 2.5% glutaraldehyde solution with SEMpore (Hitachi). The samples were lyophilized and coated with gold prior to microscopic observations. Total genomic DNA was extracted with an E.Z.N.A. Bacterial DNA Kit (Omega Bio-Tek, Doraville, GA, USA) according to the manufacturer's instructions. The 16S rRNA genes were amplified from the extracted genomic DNA using the universal primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′) as described previously (Lane, 1991Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt, E., Goodfellow, M. (Eds.). Nucleic Acid Techniques in Bacterial Systematics. Wiley, New York, pp.115–175.). Each polymerase chain reaction (PCR) mixture contained 1 × Premix Taq (Ex Taq Version, Takara Bio, Shiga, Japan), 0.4 μM each primer, and ~50 ng template DNA. The PCR amplification conditions were as follows: 95 °C for 5 min; followed by 35 cycles of 95 °C for 45 s, 55 °C for 45 s, and 72 °C for 90 s. To ensure complete elongation, a final step of 72 °C for 7 min was performed. Reactions were performed in a MyCycler Thermocycler (BioRad, Hercules, CA, USA). The PCR products were sequenced by Takara (Dalian, China). The identification of phylogenetic neighbors was initially carried out using the BLASTN program against the database containing type strains with valid published prokaryotic names, acquired from the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net) (Kim et al., 2012Kim OS, Cho YJ, Lee K et al. (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721.). Different 16S rRNA gene sequences from GenBank were aligned using CLUSTALW version 2.0 (Larkin et al., 2007Larkin MA, Blackshields G, Brown NP et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948.). A phylogenetic tree was created using the neighbor-joining method with MEGA5.0 software (Tamura et al., 2011Tamura K, Peterson D, Peterson N et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739.). A bootstrap analysis based on 1000 replicates was used to place confidence estimates on the tree.
Growth and 2,4-D-degradation conditions
Strain BJ71 was pre-cultured overnight in Luria-Bertani (LB) medium supplemented with 500 mg L−1 2,4-D. The cells were harvested by centrifugation at 6,000 g for 5 min, and then washed with sterilized MSM. For all experiments, the cell concentration of the inoculum was adjusted to an OD420 of 0.1 (corresponding to 4.6 × 107 cells mL−1) when inoculated into 150 mL MSM (pH 7.0) containing 2,4-D as the sole carbon source in a 250-mL Erlenmeyer flask. Flasks were incubated with shaking at 150 rpm on a rotary shaker. For controls, uninoculated media were maintained and tested in the same manner as described above. Culture samples were extracted at 24-h intervals to measure cell growth and the amount of 2,4-D remaining, as described below. We examined the effects of initial 2,4-D concentration (200, 350, 500, 650, and 800 mg L−1), culture temperature (22, 26, 30, and 34 °C), inoculum size (1%, 3%, 5%, 7%, and 10%), medium pH (5.0, 6.0, 7.0, 8.0, 9.0, and 10.0), and liquid medium volume (50, 100, 120, 150 mL) on cell growth and 2,4-D biodegradation. All experiments were independently performed in triplicate. Cell growth was quantified by measuring the absorbance of the sample at 420 nm using a Beckman DU640 spectrophotometer (Smejkal et al., 2001bSmejkal CW, Vallaeys T, Burton SK et al. (2001b) A rapid method to screen degradation ability in chlorophenoxyalkanoic acid herbicide-degrading bacteria. Lett Appl Microbiol 32:273–277.). The degradation rate was analyzed by determining the amount of 2,4-D remaining in the culture medium by HPLC. We used a Waters 600 HPLC system equipped with a C18 reverse-phase column with methanol − 0.1% H3PO4 (60:40) as the eluent. A decrease in the value of A230 at an elution time of ~5.5 min indicated 2,4-D metabolism. A standard curve of 2,4-D was used to calculate the percentage of degradation. Cultures were preprocessed according to the method of Suwa et al. (1996)Suwa Y, Wright AD, Fukimori F et al. (1996) Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl Environ Microbiol 62:2464–2469..
Detection of substrate degradation by strain BJ71
Strain BJ71 was precultured to an optical density of 0.6–1.0 (OD420) and then serially diluted in 1/4 Ringers solution. The 10−5 dilution of this culture was plated onto modified Loos agar plates containing (NH4)2SO4 0.1 g L−1, eosin B 0.04 g L−1, yeast extract 0.25 g L−1, Na2EDTA 0.12 g L−1, NaOH 0.02 g L−1, ZnSO4 0.004 g L−1, CuSO4 0.001 g L−1, Na2SO4 0.0001 g L−1, Na2MoO4 0.001 g L−1, CoCl2 0.0001 g L−1, MnSO4 0.0004 g L−1, 0.5 mL H2SO4, and 10 mL 1% (w/v) alkaline methylene blue solution. Separate carbon sources (MCPA, mecoprop, quizalofop, or fluroxypyr) were added at concentrations of 3 mmol L−1. Plates were set up in triplicate and incubated for 4 days at 25 °C. This method was described by Smejkal et al. (2001bSmejkal CW, Vallaeys T, Burton SK et al. (2001b) A rapid method to screen degradation ability in chlorophenoxyalkanoic acid herbicide-degrading bacteria. Lett Appl Microbiol 32:273–277.; 2003)Smejkal CW, Seymour FA, Burton SK et al. (2003) Characterisation of bacterial cultures enriched on the chlorophenoxyalkanoic acid herbicides 4-(2,4-dichlorophenoxy) butyric acid and 4-(4-chloro-2-methylphenoxy) butyric acid. J Ind Microbiol Biotechnol 30:561–567..
Soil microcosm degradation experiments
Organic agricultural topsoil, classified as a loamy soil with 14% clay, 32% silt, 54% sand, and 2.34% carbon (pH 6.9) was collected from a farm (N 26°25′59″ E 106°40′16″) (Guizhou, China), and then sieved (pore size, 2 mm) and air dried. The soil had not been exposed to 2,4-D previously. Soil (200 g) was added to 500-mL wide-mouth glass jars covered with aluminum foil. Control soils were sterilized by autoclaving (121 °C, 20 min) on 3 different days before starting the microcosm experiments. The soil used in the microcosm experiments was supplemented with 2,4-D at a final concentration of 350 mg kg−1. Strain BJ71 was grown at 30 °C in LB with 2,4-D (500 mg L−1), harvested by centrifugation, washed twice, and then resuspended in sodium phosphate buffer. The soil samples were incubated for 1 day to equilibrate, and then inoculated with freshly grown cultures at a cell density of 7 × 107 cfu g−1 soil. Sterilized controls containing the same amount of 2,4-D were run simultaneously under identical conditions. After all amendments were added, the moisture content of soil was adjusted with water to 75% of the water-holding capacity. The soil was shaken every week to enhance O2 availability. Soil microcosms were incubated at 30 °C in the dark to prevent photodegradation of 2,4-D during the 42-day experiment. All experiments were conducted with three replicates. The concentration of 2,4-D in the soils was determined by HPLC according to the method of Holben et al. (1992)Holben WE, Schroeter BM, Calabrese VGM et al. (1992) Gene probe analysis of soil microbial populations selected by amended with 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol 58:3941–3948..
Statistical analysis
Data were analyzed using one-way analysis of variance and multiple comparisons were performed with Duncan's multiple range test using SPSS software for Windows (version 19.0).
Results
Isolation and screening of 2,4-D-degrading bacterial strains
We isolated 22 strains of 2,4-D-degrading bacteria by picking colonies with differing morphologies. All the strains were able to utilize 2,4-D as the sole carbon and energy source. Of the 22 strains, 13 degraded more than 60% of the initial 2,4-D (500 mg L−1) during 1 week of incubation at 30 °C. Sequence analyses showed that the genomes of these strains contained the conserved sequence of a class I tfdA gene (Han et al., 2014Han L, Liu Y, He A et al. (2014) 16S rRNA gene phylogey and tfdA gene analysis of 2,4-D-degrading bacteria isolated in China. World J Microbiol Biotechnol. Doi: 10.1007/s11274-014-1680-6.
https://doi.org/10.1007/s11274-014-1680-...
). The BJ71 strain accumulated the largest biomass and showed the most complete degradation of 2,4-D among these 2,4-D-degrading strains. Therefore, BJ71 was selected for further investigation.
Identification of strain BJ71
According to the observation of cell morphology and physiological and biochemicala tests, BJ71 is a motile, Gram-negative rod-shaped bacterium that forms opalescent colonies (Figure 1) with oxidase and catalase activities. The bacterium showed positive results in urease and nitrate reduction tests, but negative results in tests for glucose fermentation and indole and citrate utilization. The 16S rRNA sequence of strain BJ71 (1,390 bp) was compared with bacterial 16S rRNA sequences in GenBank. Phylogenetic neighbors were identified by BLASTN searches against the database containing type strains with valid published prokaryotic names, acquired from the EzTaxon-e server. The 16S rRNA gene sequence of strain BJ71 showed 100% similarity to that of Cupriavidus campinensis WS2T (GenBank accession nos. AF312020) (Figure 2). C. campinensis is a new species assigned in 2001. A metal-resistant type strain, WS2T, was isolated from a zinc-desertified area in Belgium (Goris et al., 2001Goris J, De Vos P, Coenye T et al. (2001) Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al., 1998 emend. Int J Syst Evol Microbiol 51:1773–1782.).
Colonies and cells of strain BJ71. (A) Photograph of BJ71 colonies on LB agar plate (B) Scanning electron micrograph of BJ71 cells.
NJ tree based on 16S rRNA gene (1,390 bp) sequences showing the phylogenetic relationship between strain BJ71 and other related species in genera Ralstonia and Wautersia sp. GenBank accession number are in parentheses. Bootstrap values are based on 1,000 resampled datasets. All major branches were conserved in the maximum-parsimony tree. Burkholderia andropogoni ATCC 23061T (X67037) served as the outgroup. Scale bar indicates number of nucleotide changes per base position analyzed.
Based on its 16S rRNA gene sequence analysis and its morphological, physiological, and biochemical characteristics, strain BJ71 was identified as Cupriavidus campinensis. The nucleotide sequence of the 16S rRNA gene of strain BJ71 has been deposited in GenBank under the accession number KF997830.
Effect of environmental factors on growth and 2,4-D degradation of strain BJ71
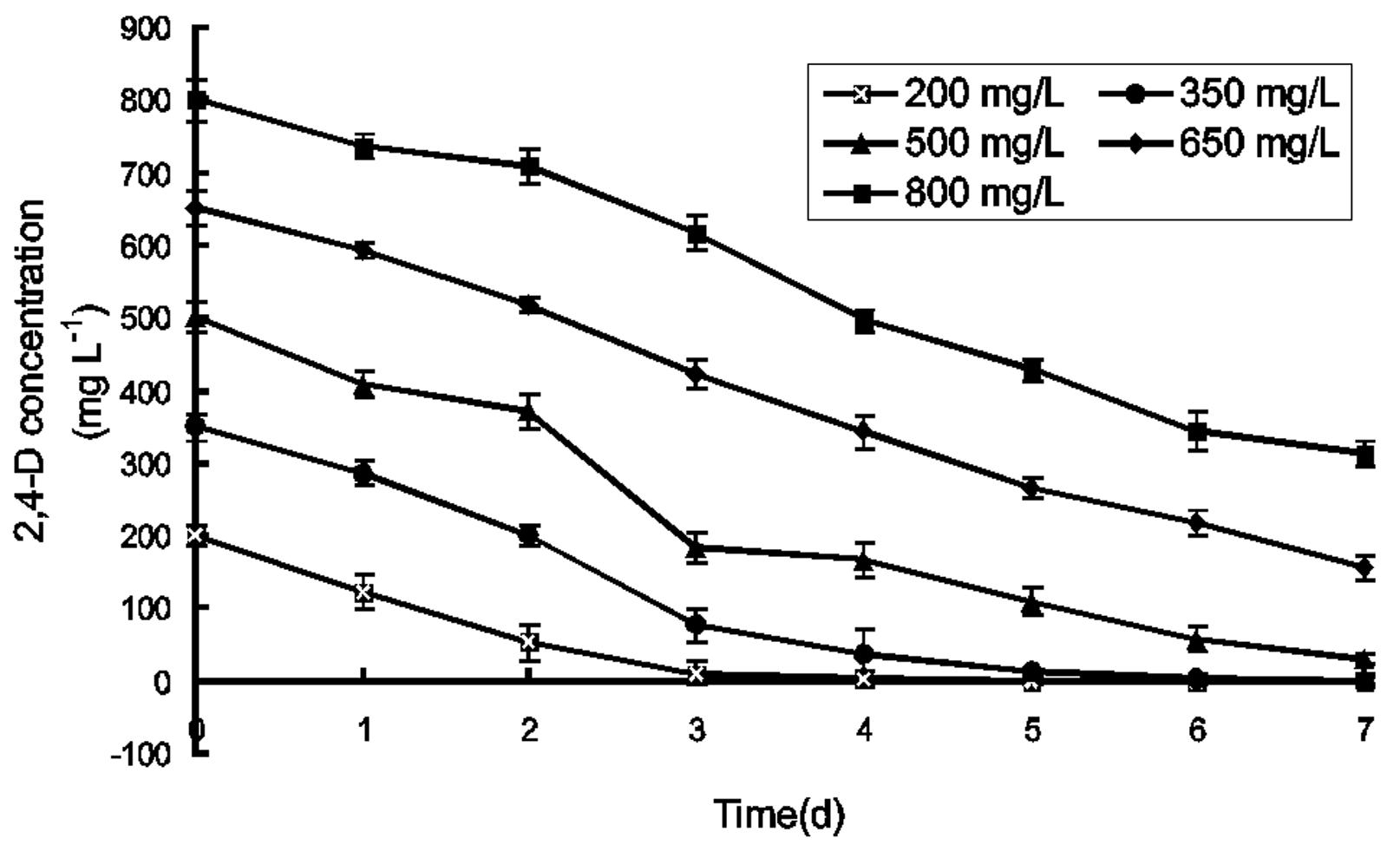
We investigated the effect of different initial 2,4-D concentrations on 2,4-D degradation by strain BJ71 (Figure 3). Strain BJ71 degraded 2,4-D quickly at low substrate concentrations (200 and 350 mg L−1), and showed almost complete degradation of 200 mg L−1 2,4-D in 3 days. When the initial 2,4-D concentration was 350 mg L−1, BJ71 degraded 90% of the 2,4-D after 4 days. BJ71 was also able to degrade 2,4-D at high concentrations in MSM (94.08% of 500 mg L−1 and 61% of 800 mg L−1 after 7 days of incubation). We also investigated the effects of other factors on the 2,4-D degradation rate of strain BJ71 (Figure 4). Among six different pHs, pH 7.0 was optimal for 2,4-D degradation by BJ71. This strain was well-adapted to a wide range of pHs and was able to degrade up to 40% of the 2,4-D at pH 5.0–10.0. Temperature is a major environmental factor affecting degradation. The biodegradation rate was approximately 8.49% at the highest temperature tested (34 °C), and 98.98% at 30 °C. The optimum inoculum size was 3.0%. The degradation rate decreased when the inoculum size was too low or too high because of insufficient inoculum or nutrient limitation, respectively. There was no significant effect of different medium volumes on the 2,4-D degradation rate.
Effect of initial 2,4-D concentration on degradation of 2,4-D by strain BJ71. Incubation conditions were as follows: pH 7.0, final cell density at 420 nm adjusted to 0.1. Cells were inoculated into 150 mL MSM (pH 7.0) in a 250-mL flask at 30 °C and grown with shaking at 150 rpm on a rotary shaker. Standard errors are within 5% of the mean.
Effects of environmental conditions on degradation rate of 2,4-D by strain BJ71. Effects of pH (a), temperature (b), inoculum size (c), and medium volume (d) on 2,4-D degradation. For all experiments, final density of cells at 420 nm was adjusted to 0.1. Cells were inoculated into 150 mL MSM (pH 7.0) containing 350 mg L−1 2,4-D in a 250-mL Erlenmeyer flask before incubation at 30 °C with shaking at 150 rpm, unless otherwise stated. All data were obtained at 120 h of incubation. Values shown are means of duplicate experiments. Error bars indicate standard deviation (SD). * indicates significant difference (Duncan's test, p < 0.05).
The optimal conditions for 2,4-D biodegradation by strain BJ71 were as follows: initial pH of 7.0, incubation temperature of 30 °C, inoculum size of 3.0% (v/v), and an initial 2,4-D concentration of 350 mg L−1. The typical growth of strain BJ71 was consistent with the degradation curves under optimal conditions (Figure 5). Up to 99.57% of the 2,4-D was degraded by strain BJ71 under these optimum conditions after 6 days of incubation.
Amount of 2,4-D remaining and cell growth during degradation of 2,4-D by strain BJ71. Incubation conditions were as follows: pH 7.0, final cell density at 420 nm adjusted to 0.1. Cells were inoculated into 150 mL MSM (pH 7.0) containing 350 mg L−1 2,4-D in a 250-mL Erlenmeyer flask at 30 °C and shaker at 150 rpm. Standard errors are within 5% of the mean.
Analysis of substrate range
It has been reported that growth on Modified Loos medium can reveal microbial dehalogenation of herbicide compounds, during which there is a cleavage of the aromatic ring (Smejkal et al., 2003Smejkal CW, Seymour FA, Burton SK et al. (2003) Characterisation of bacterial cultures enriched on the chlorophenoxyalkanoic acid herbicides 4-(2,4-dichlorophenoxy) butyric acid and 4-(4-chloro-2-methylphenoxy) butyric acid. J Ind Microbiol Biotechnol 30:561–567.). BJ71 was grown on this medium supplemented with 3 mmol of several other herbicides including mecoprop, MCPA, quizalofop, and fluroxypyr. After 4 days of incubation, dark violet colonies were observed on the plates of Modified Loos medium containing 2,4-D, quizalofop and fluroxypyr. Therefore, BJ71 was able to grow on media containing the herbicides quizalofop and fluroxypyr, but not on media containing MCPA or mecoprop.
Degradation of 2,4-D in soil
We evaluated the patterns of degradation in non-sterilized and sterilized soils, with and without strain BJ71 (Figure 6). In non-sterile soil inoculated with strain BJ71, the degradation rate was markedly higher than in the other treatments after 42 days of incubation at 30 °C. After 14 days of incubation, 87.13% and 42.53% of 2,4-D (initial concentration of 350 mg kg−1) were degraded in non-sterile and sterilized soil inoculated with the strain BJ71, respectively. In non-sterile control without strain BJ71, the removal rate was only 7.34%. There was no degradation of 2,4-D in control sterilized soil, which contained 350 mg kg−1 2,4-D but no inoculant (data not shown). The decrease in 2,4-D concentrations in soil lacking BJ71 indicated that other indigenous microorganisms showed some 2,4-D degrading ability. Strain BJ71 enhanced the degradation of 2,4-D when present with other indigenous microorganisms in the soil. The isolated C. campinensis BJ71 strain has been deposited in the China Centre for Type Culture Collection (Accession number CCTCC M 2014006).
Degradation of 350 mg kg−1 2,4-D in soil inoculated with strain BJ71. Soil in microcosm degradation experiments contained 2,4-D at 350 mg L−1, and was inoculated with BJ71 at a cell density of 7 × 107 cfu g−1 soil. Soil microcosms were incubated at 30 °C for 42 days in darkness to prevent photodegradation of 2,4-D. Values shown are the mean of three replicates. Standard errors are within 5% of mean value.
Discussion
In this study, we isolated an indigenous 2,4-D degrading bacterium from a polluted wheat field in Beijing, China. The strain was identified as C. campinensis based on its 16S rRNA gene sequence analysis and its physiological features. In previous studies, several types of 2,4-D-degrading bacteria were isolated from 2,4-D-treated environments. These bacteria have been categorized into three groups based on their degradation enzymes and physiological properties. The first group consists of copiotrophic bacteria belonging to the β and γ subdivisions of the class Proteobacteria, and includes species in the genera Achromobacter, Burkholderia, Delftia, Halomonas, Pseudomonas, Ralstonia, Rhodoferax, and Variovorax. Their 2,4-D-catabolizing enzymes are encoded by genes homologous to tfdA, which encodes an Fe(II)/α-ketoglutarate-dependent dioxygenase that converts 2,4-D into 2,4-dichlorophenol. The tfdA gene is initially found in R. eutropha JMP134 (Gonzalez et al., 201; Lee et al., 2005Lee T, Kurata S, Nakatsu C et al. (2005) Molecular analysis of bacterial community based on 16S rDNA and functional genes in activated sludge enriched with 2,4-dichlorophenoxyacetic acid (2,4-D) under different cultural conditions. Microb Ecol 49:151–162., Vallaeys et al., 1999Vallaeys T, Courde L, McGowan C et al. (1999) Phylogenetic analyses indicate independent recruitment of diverse gene cassettes during assemblage of the 2,4-D catabolic pathway. FEMS Microbiol Ecol 28:373–382.). The second group consists of members of the α-Proteobacteria, closely related to Bradyrhizobium sp. (Itoh et al., 2000Itoh K, Kanda R, Momoda Y et al. (2000) Presence of 2,4-D-catabolizing bacteria in a Japanese arable soil that belong to BANA (Bradyrhizobium-Agromonas-Nitrobacter-Afipia) cluster in α-Proteobacteria. Microbes Environ 15:113–117.; Kamagata et al., 1997Kamagata Y, Fulthorpe RR, Tamura K et al. (1997) Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol 63:2266–2272.). Members of the third group are copiotrophic α-Proteobacteria in the genus Sphingomonas (McGowan et al., 1998McGowan C, Fulthorpe R, Wright A et al. (1998) Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl Environ Microbiol 64:4089–4092.; Suwa et al., 1996Suwa Y, Wright AD, Fukimori F et al. (1996) Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl Environ Microbiol 62:2464–2469.). These strains, which include Delftia acidovorans MC1, Rhodoferax sp. P230, and Sphingobium herbicidovorans MH, contain sdpA or rdpA genes, which encode products that can cleave enantiomers of racemic compounds (Mller et al., 2006Mller TA, Fleischmann T, van der Meer JR et al. (2006) Purification and characterization of two enantioselective α-ketoglutarate-dependent dioxygenases, RdpA and SdpA, from Sphingomonas herbicidovorans MH. Appl Environ Microbiol 72:4853–4861.; Schleinitz et al., 2004Schleinitz KM, Kleinsteuber S, Vallaeys T et al. (2004) Localization and characterization of two novel genes encoding stereospecific dioxygenases catalyzing 2(2,4-dichlorophenoxy)propionate cleavage in Delftia acidovorans MC1. Appl Environ Microbiol 70:5357–5365.; Westendorf et al., 2006Westendorf A, Benndorf D, Pribyl T et al. (2006) Kinetic traits and enzyme form patterns of (R)-2-(2,4-dichlorophenoxy)propionate/α-ketoglutarate dioxygenase (RdpA) after expression in different bacterial strains. Eng Life Sci 6:552–559.). These 2,4-D-degrading isolates have been well characterized in previous studies. The degradation capacity of strain BJ71 was greater than those reported for Sphingomonas agrestis strain 58-1 (Shimojo et al., 2009Shimojo M, Kawakami M, Amada K (2009) Analysis of genes encoding the 2,4-dichlorophenoxyacetic acid-degrading enzyme from Sphingomonas agrestis 58-1. J Biosci Bioeng 108:56–59.) and three other strains; Burkholderia cepacia DS-1, Pseudomonas sp. DS-2, and Sphingomonas paucimobilis DS-3 (Cycon et al., 2011Cycon M, Zmijowska A, Piotrowska-Seget Z (2011) Biodegradation kinetics of 2,4-D by bacterial strains isolated from soil. Cent Eur J Biol 6:188–198.). The 2,4-D degradation ability of strain BJ71 in medium containing 2,4-D was comparable to those reported for C. pampae CPDB6T (Cuadrado et al., 2010Cuadrado V, Gomila M, Merini L et al. (2010) Cupriavidus pampae sp. nov., a novel herbicide-degrading bacterium isolated from agricultural soil. Int J Syst Evol Microbiol 60:2606–2612.) and Halomonadaceae sp. I-18 (Maltseva et al., 1996Maltseva O, McGowan C, Fulthorpe R et al. (1996) Degradation of 2,4-dichlorophenoxyacetic acid by haloalkaliphilic bacteria. Microbiology 142:1115–1122.). The 2,4-D degradation efficiency of strain BJ71 was greater than that of C. necator JMP134, the most intensively studied chloroaromatic-degrading microorganism to date. When grown on medium containing 250 mg L−1 of 2,4-D as the sole carbon and energy source, JMP134 showed a maximum 2,4-D degradation rate of 70% after 10 days of incubation (Lerch et al., 2007Lerch TZ, Dignac MF, Barriuso E et al. (2007) Tracing 2,4-D metabolism in Cupriavidus necator JMP134 with 13C-labelling technique and fatty acid profiling. J Microbiol Methods 71:162–174.). A Delftia species isolated from a polluted river in Argentina was able to degrade 200 mg L−1 of 2,4-D in 28 h and remove 99.0% of the pollutant, whereas C. necator EMA-G isolated from agricultural soil in Argentina degraded 250 mg L−1 2,4-D in less than 1 day (Gonzale et al., 2012Gonzalez AJ, Gallego A, Gemini VL et al. (2012) Degradation and detoxification of the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) by an indigenous Delftia sp. strain in batch and continuous systems. Int Biodeterior Biodegradation 66:8–13.; Zabaloy and Gómez, 2013Zabaloy MC, Gómez MA (2013) Isolation and characterization of indigenous 2,4-D herbicide degrading bacteria from an agricultural soil in proximity of Sauce Grande River, Argentina. Ann Microbiol doi 10.1007/s13213-013-0731-9.
https://doi.org/10.1007/s13213-013-0731-...
). B. cepacia YK-2 and a strain of P. putida isolated from the Jordan Valley completely degraded 500 mg L−1 2,4-D within 28 and 45 h, respectively (Cho et al., 2002Cho YS, Kahng HY, Kim CK et al. (2002) Physiological and cellular response of the 2,4-D degrading bacterium, Burkholderia cepacia YK-2, to the phenoxyherbicides 2,4-D and 2,4,5-T. Curr Microbiol 45:415–422.; Khalil, 2003Khalil AB (2003) Isolation and characterization of 2,4-dichlorophenoxyacetic acid degrading organisms from soil in Jordan Valley. Biotechnology 2:73–85.). The 2,4-D degradation efficiency of these strains in shaking culture with higher concentrations of 2,4-D or in soil microcosms was not reported. In this study, BJ71 in liquid culture showed a degradation rate of up to 60% after 7 days of incubation when grown on medium containing 800 mg L−1 2,4-D.
Many 2,4-D-degrading strains do not have wide substrate specificity for chlorophenoxyalkanoic herbicides, and are limited to 2,4-D as a growth substrate (Hoffman et al., 1996Hoffman D, Mller RH, Kiesel B et al. (1996) Isolation and characterization of an alkaliphilic bacterium capable of growing on 2,4-dichlorophenoxyacetic acid and 4-chloro-2-methylphenoxyacetic acid. Acta Biotechnologica 16:121–131.; Ka et al., 1994Ka JO, Holben WE, Tiedje JM (1994) Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol 60:1106–1115.). We screened an expanded set of structurally related herbicides, and unexpectedly found that this 2,4-D-degrading C. campinensis BJ71 strain could also degrade quizalofop and fluroxypyr. The mechanism of action of these herbicides is entirely different from that of 2,4-D. Quizalofop is in the aryloxyphenoxypropionate (AOPP) class of potent grass-selective herbicides, and fluroxypyr consists of pyridyloxyacetate compounds. Wright et al. (2010)Wright TR, Shan G, Walsh TA et al. (2010) Robust crop resistance to broadleaf and grass herbicides provided by aryloxyalkanoate dioxygenase transgenes. PNAS 107:20240–20245. reported that AAD-1 (synonymous with RdpA in S. herbicidivorans) has the unique ability to enantioselectively cleave R-quizalofop, and that AAD-12, (synonymous with SdpA from D. acidovorans) can degrade fluroxypyr. However, there have been no other reports in the literature of 2,4-D-degraders containing the tfdA gene that are able to degrade fluroxypyr and quizalofop. Recently, we cloned the entire tfdA gene of C. campinensis strain BJ71 (GenBank accession number: KJ028765). The sequence of the tfdA gene from C. campinensis BJ71 shows some differences from that of C. necator JMP134 (Han et al., 2015Han L, Liu Y, Li C et al. (2015) Cloning, expression, characterization and mutational analysis of the tfdA gene from Cupriavidus campinensis BJ71. World J Microbiol Biotechnol. Doi 10.1007/s11274-015-1852-z.
https://doi.org/10.1007/s11274-015-1852-...
). To our knowledge, this is the first report of a 2,4-D degrader containing the tfdA that can degrade several different synthetic auxins (2,4-D, fluroxypyr, and the aryloxyphenoxypropionate herbicide quizalofop). Because of these characteristics, this strain has great potential for bioremediation of these herbicides in polluted environments.
For successful bioremediation, it is important to identify and isolate appropriate microbial strains, and to ensure their survival and activity after their release into the target habitat. A key factor in the failure of bioaugmentation strategies is the rapid decline of the population of introduced cells. The introduced strain may face intense competition, predation, or parasitism in the natural environment. The most suitable candidate for bioaugmentation is an autochthonous strain that is able to survive long-term in the target habitat, and continuously degrade the contaminant (Ramos et al., 1991Ramos JL, Duque E, Ramos-Gonzalez MI (1991) Survival in soils of an herbicide-resistant Pseudomonas putida strain bearing a recombinant TOL plasmid. Appl Environ Microbiol 57:260–266.; Thompson et al., 2005Thompson I, van der Gast CJ, Ciric L et al. (2005) Bioaugmentation for bioremediation: the challenge of strain selection. Environ Microbiol 7:909–915.). It has been reported that the batch culture method used to enrich and isolate bacteria is highly selective for fast-growing copiotrophic microbes, and that the isolation of Cupriavidus sp. from the most dilute positive MPN tubes may reflect its dominance in the 2,4-D degrading community in soil (Macur et al., 2007Macur RE, Wheeler JT, Burr MD et al. (2007) Impacts of 2,4-D application on soil microbial community and on populations associated with 2,4-D degradation. Microbiol Res 162:37–45.; Zabaloy and Gómez, 2013Zabaloy MC, Gómez MA (2013) Isolation and characterization of indigenous 2,4-D herbicide degrading bacteria from an agricultural soil in proximity of Sauce Grande River, Argentina. Ann Microbiol doi 10.1007/s13213-013-0731-9.
https://doi.org/10.1007/s13213-013-0731-...
). Our results show that Cupriavidus sp. BJ71, isolated from a polluted wheat field in China, can rapidly degrade 2,4-D in liquid medium. The biodegradation of 2,4-D was much more rapid in 2,4-D (350 mg kg−1)-treated soil inoculated with BJ71 than in soil without this strain. These results suggest that C. campinensis BJ71 could be a suitable candidate for bioaugmentation for in situ clean-up strategies. Because it is an indigenous organism, it is more likely to survive than are other non-indigenous bacteria when introduced into polluted soil and water environments in China.
Acknowledgments
This work was supported by the Foundation of the National Transgenic Major Project of China (2013ZX08010-003, 2014ZX0801008B-002) and the Province Science Project in Guizhou (J [2013] 2123).
References
- Chinalia FA, Killham KS (2006) 2,4-Dichlorophenoxyacetic acid (2,4-D) biodegradation in river sediments of northeast-Scotland and its effect on the microbial communities (PLFA and DGGE). Chemosphere 64:1675–1683.
- Chinalia FA, Regali-Seleghin MH, Correa EM (2007) 2,4-D toxicity: cause, effect and control. Terrestrial and Aquatic Environmental Toxicology 1:24–33.
- Cho YS, Kahng HY, Kim CK et al. (2002) Physiological and cellular response of the 2,4-D degrading bacterium, Burkholderia cepacia YK-2, to the phenoxyherbicides 2,4-D and 2,4,5-T. Curr Microbiol 45:415–422.
- Cuadrado V, Gomila M, Merini L et al. (2010) Cupriavidus pampae sp. nov., a novel herbicide-degrading bacterium isolated from agricultural soil. Int J Syst Evol Microbiol 60:2606–2612.
- Cycon M, Zmijowska A, Piotrowska-Seget Z (2011) Biodegradation kinetics of 2,4-D by bacterial strains isolated from soil. Cent Eur J Biol 6:188–198.
- El-Bestawy E, Hans-Jorgen A (2007) Effect of nutrient amendments and sterilization on mineralization and/or biodegradation of 14C-labeled MCPP by soil bacteria under aerobic conditions. Int Biodeterior Biodegradation 59:193–201.
- Fukumori F, Hausinger RP (1993a) Alcaligenes eutrophus JMP134 "2,4-dichlorophenoxyacetate monooxygenase" is an α-ketoglutarate-dependent dioxygenase. J Bacteriol 175:2083–2086.
- Fukumori F, Hausinger RP (1993b) Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J Biol Chem 268:24311–24317.
- Garc'a GB, Konjuh C, Duffard RO et al. (2006) Dopamine-β-hydroxylase immunohistochemical study in the locus coeruleus of neonate rats exposed to 2,4-dichlorophenoxyacetic acid through mother's milk. Drug Chem Toxicol 29:435–442.
- Garrity GM, Bell JA, Lilburn TG (2004) Taxonomic outline of the prokaryotes Bergey's Manual of Systematic Bacteriology (second edition) Springer New York Berlin Heidelberg. doi: 10.1007/bergeysoutline 200405.
» https://doi.org/10.1007/bergeysoutline200405 - Gonzalez AJ, Gallego A, Gemini VL et al. (2012) Degradation and detoxification of the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) by an indigenous Delftia sp. strain in batch and continuous systems. Int Biodeterior Biodegradation 66:8–13.
- Goris J, De Vos P, Coenye T et al. (2001) Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al., 1998 emend. Int J Syst Evol Microbiol 51:1773–1782.
- Han L, Liu Y, He A et al. (2014) 16S rRNA gene phylogey and tfdA gene analysis of 2,4-D-degrading bacteria isolated in China. World J Microbiol Biotechnol. Doi: 10.1007/s11274-014-1680-6.
» https://doi.org/10.1007/s11274-014-1680-6 - Han L, Liu Y, Li C et al. (2015) Cloning, expression, characterization and mutational analysis of the tfdA gene from Cupriavidus campinensis BJ71. World J Microbiol Biotechnol. Doi 10.1007/s11274-015-1852-z.
» https://doi.org/10.1007/s11274-015-1852-z - Hoffman D, Mller RH, Kiesel B et al. (1996) Isolation and characterization of an alkaliphilic bacterium capable of growing on 2,4-dichlorophenoxyacetic acid and 4-chloro-2-methylphenoxyacetic acid. Acta Biotechnologica 16:121–131.
- Holben WE, Schroeter BM, Calabrese VGM et al. (1992) Gene probe analysis of soil microbial populations selected by amended with 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol 58:3941–3948.
- Itoh K, Kanda R, Momoda Y et al. (2000) Presence of 2,4-D-catabolizing bacteria in a Japanese arable soil that belong to BANA (Bradyrhizobium-Agromonas-Nitrobacter-Afipia) cluster in α-Proteobacteria. Microbes Environ 15:113–117.
- Ka JO, Holben WE, Tiedje JM (1994) Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl Environ Microbiol 60:1106–1115.
- Kamagata Y, Fulthorpe RR, Tamura K et al. (1997) Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol 63:2266–2272.
- Khalil AB (2003) Isolation and characterization of 2,4-dichlorophenoxyacetic acid degrading organisms from soil in Jordan Valley. Biotechnology 2:73–85.
- Kim OS, Cho YJ, Lee K et al. (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721.
- Laganá A, Bacaloni A, De Leva I et al. (2002) Occurrence and determination of herbicides and their major transformation products in environmental waters. Anal Chim Acta 462:187–198.
- Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt, E., Goodfellow, M. (Eds.). Nucleic Acid Techniques in Bacterial Systematics. Wiley, New York, pp.115–175.
- Larkin MA, Blackshields G, Brown NP et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948.
- Lee T, Kurata S, Nakatsu C et al. (2005) Molecular analysis of bacterial community based on 16S rDNA and functional genes in activated sludge enriched with 2,4-dichlorophenoxyacetic acid (2,4-D) under different cultural conditions. Microb Ecol 49:151–162.
- Lerch TZ, Dignac MF, Barriuso E et al. (2007) Tracing 2,4-D metabolism in Cupriavidus necator JMP134 with 13C-labelling technique and fatty acid profiling. J Microbiol Methods 71:162–174.
- Macur RE, Wheeler JT, Burr MD et al. (2007) Impacts of 2,4-D application on soil microbial community and on populations associated with 2,4-D degradation. Microbiol Res 162:37–45.
- Maloney EK, Waxman DJ (1999) Trans-activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol Appl Pharmacol 161:209–218.
- Maltseva O, McGowan C, Fulthorpe R et al. (1996) Degradation of 2,4-dichlorophenoxyacetic acid by haloalkaliphilic bacteria. Microbiology 142:1115–1122.
- Manzano M, Morán AC, Tesser B et al. (2007) Role of eukaryotic microbiota in soil survival and catabolic performance of the 2,4-D herbicide degrading bacteria Cupriavidus necator JMP134. Antonie Van Leeuwenhoek 91:115–126.
- Marrón-Montiel E, Ordaz-Ruiz N, Rubio-Granados C et al. (2006) 2,4-D-degrading bacterial comsortium isolation kinetic characterization in batch and continuous culture and application for bioaugmenting an activated sludge microbial community. Process Biochem 41:1521–1528.
- McGowan C, Fulthorpe R, Wright A et al. (1998) Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl Environ Microbiol 64:4089–4092.
- Mller TA, Fleischmann T, van der Meer JR et al. (2006) Purification and characterization of two enantioselective α-ketoglutarate-dependent dioxygenases, RdpA and SdpA, from Sphingomonas herbicidovorans MH. Appl Environ Microbiol 72:4853–4861.
- Ramos JL, Duque E, Ramos-Gonzalez MI (1991) Survival in soils of an herbicide-resistant Pseudomonas putida strain bearing a recombinant TOL plasmid. Appl Environ Microbiol 57:260–266.
- Sánchez MA, Vásquez M, González B (2004) A previously unexposed forest soil microbial community degrades high levels of the pollutant 2,4,6-trichlorophenol. Appl Environ Microbiol 70:7567–7570.
- Schleinitz KM, Kleinsteuber S, Vallaeys T et al. (2004) Localization and characterization of two novel genes encoding stereospecific dioxygenases catalyzing 2(2,4-dichlorophenoxy)propionate cleavage in Delftia acidovorans MC1. Appl Environ Microbiol 70:5357–5365.
- Shimojo M, Kawakami M, Amada K (2009) Analysis of genes encoding the 2,4-dichlorophenoxyacetic acid-degrading enzyme from Sphingomonas agrestis 58-1. J Biosci Bioeng 108:56–59.
- Singh DK (2008) Biodegradation and bioremediation of pesticide in soil: concept, method and recent developments. Indian J Microbiol 48:35–40.
- Smejkal CW, Vallaeys T, Seymour FA et al. (2001a) Characterization of (R/S)-mecoprop [2-(2-methyl-4-chlorophenoxy)propionic acid]-degrading Alcaligenes sp. CS1 and Ralstonia sp. CS2 isolated from agricultural soils. Environ Microbiol 3:288–293.
- Smejkal CW, Vallaeys T, Burton SK et al. (2001b) A rapid method to screen degradation ability in chlorophenoxyalkanoic acid herbicide-degrading bacteria. Lett Appl Microbiol 32:273–277.
- Smejkal CW, Seymour FA, Burton SK et al. (2003) Characterisation of bacterial cultures enriched on the chlorophenoxyalkanoic acid herbicides 4-(2,4-dichlorophenoxy) butyric acid and 4-(4-chloro-2-methylphenoxy) butyric acid. J Ind Microbiol Biotechnol 30:561–567.
- Streber WR, Timmis KN, Zenk MH (1987) Analysis, cloning, and high-level expressions of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol 169:2950–2955.
- Suwa Y, Wright AD, Fukimori F et al. (1996) Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl Environ Microbiol 62:2464–2469.
- Tamura K, Peterson D, Peterson N et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739.
- Thompson I, van der Gast CJ, Ciric L et al. (2005) Bioaugmentation for bioremediation: the challenge of strain selection. Environ Microbiol 7:909–915.
- Vallaeys T, Courde L, McGowan C et al. (1999) Phylogenetic analyses indicate independent recruitment of diverse gene cassettes during assemblage of the 2,4-D catabolic pathway. FEMS Microbiol Ecol 28:373–382.
- Vandamme P, Coenye T (2004) Taxonomy of the genus Cupriavidus: a tale of lost and found. Int J Syst Evol Microbiol 54:2285–2289.
- Vaneechoutte M, K?mpfer P, De Baere T et al. (2004) Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al., 1990) comb. nov. Int J Syst Evol Microbiol 54:317–327.
- Westendorf A, Benndorf D, Pribyl T et al. (2006) Kinetic traits and enzyme form patterns of (R)-2-(2,4-dichlorophenoxy)propionate/α-ketoglutarate dioxygenase (RdpA) after expression in different bacterial strains. Eng Life Sci 6:552–559.
- Wright TR, Shan G, Walsh TA et al. (2010) Robust crop resistance to broadleaf and grass herbicides provided by aryloxyalkanoate dioxygenase transgenes. PNAS 107:20240–20245.
- Zabaloy MC, Gómez MA (2013) Isolation and characterization of indigenous 2,4-D herbicide degrading bacteria from an agricultural soil in proximity of Sauce Grande River, Argentina. Ann Microbiol doi 10.1007/s13213-013-0731-9.
» https://doi.org/10.1007/s13213-013-0731-9
Publication Dates
-
Publication in this collection
Apr-Jun 2015
History
-
Received
09 Mar 2014 -
Accepted
30 Oct 2014