Abstract
Leptospires are usually classified by methods based on DNA-DNA hybridization and the conventional cross-agglutination absorption test, which uses polyclonal antibodies against lipopolysaccharides. In this study, the amplification of the rpoB gene, which encodes the beta-subunit of RNA polymerase, was used as an alternative tool to identify Leptospira. DNA extracts from sixty-eight serovars were obtained, and the hypervariable region located between 1990 and 2500-bp in the rpoB gene was amplified by polymerase chain reaction (PCR). The 600-bp amplicons of the rpoB gene were digested with the restriction endonucleases TaqI, Tru1I, Sau3AI and MslI, and the restriction fragments were separated by 6% polyacrylamide gel electrophoresis. Thirty-five fragment patters were obtained from the combined data of restriction fragment length polymorphism (PCR-RFLP) analysis and used to infer the phylogenetic relationships among the Leptospira species and serovars. The species assignments obtained were in full agreement with the established taxonomic classifications. Twenty-two serovars were effectively identified based on differences in their molecular profiles. However, the other 46 serovars remained clustered in groups that included more than one serovar of different species. This study demonstrates the value of RFLP analysis of PCR-amplified rpoB as an initial method for identifying Leptospira species and serovars.
Leptospira ; rpoB gene; RFLP; serovar; DNA typing
Introduction
Leptospirosis is a zoonotic disease of global importance that has emerged as a major cause of morbidity and mortality among impoverished populations (Ko et al., 2009Ko AI, Goarant C, Picardeau M (2009) Leptospira: The Dawn of the Molecular Genetics Era for an Emerging Zoonotic Pathogen. Nat Rev Microbiol 10:736–747.). Based on global data, more than 500,000 new cases of leptospirosis are reported annually, with mortality rates exceeding 10% (WHO, 1999World Health Organization. Leptospirosis worldwide. (1999). Weekly Epidemiol Rec 74:237–242., 2006World Health Organization (2006) Informal Consultation on Global Burden of Leptospirosis: Methods of Assessment. Available at: http://www.who.int/entity/foodsafety/zoonoses/InformalConsultationOnBoDLeptospirosis.pdf.
http://www.who.int/entity/foodsafety/zoo...
). Multiple factors, including environmental, demographic, social, and economic factors, have contributed to the emergence of this disease, which affects a broad range of mammalian hosts, including humans, wildlife, and domestic animals (Bharti and Nally, 2003Bharti AR, Nally JE (2003) Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3:757–771.; Lau et al., 2012Lau C, Clements A, Skelly C et al. (2012) Leptospirosis in American Samoa-Estimating and mapping risk using environmental data. PLoS Negl Trop Dis 6:e1669.).
The precise identification and classification of Leptospira spp. is important for epidemiological and public health surveillance (Mohammed et al., 2011Mohammed H, Nozha C, Hakim K et al. (2011) Leptospira: Morphology, Classification and Pathogenesis. Bacteriol Parasitol 2:6.). Leptospires are usually classified by methods based on DNA-DNA hybridization, whereas the cross-agglutination absorption test (CAAT), which uses polyclonal antibodies against lipopolysaccharides (LPSs), has led to the definition of serovars that are today considered to be the basic systematic units of Leptospira spp. (Cerqueira and Picardeau, 2009Cerqueira G, Picardeau M (2009) A century of Leptospira strain typing. Infect Genet Evol 9:760–768.; Galloway and Levett, 2010Galloway RL, Levett PN (2010) Application and Validation of PFGE for Serovar Identification of Leptospira Clinical Isolates. PLoS Negl Trop Dis 14:e824.). Serological methods for the characterization of Leptospira species are complex and costly, restricting their worldwide distribution and use (Ahmed et al., 2010Ahmed A, Anthony RM, Hartskeerl RA (2010) A simple and rapid molecular method for Leptospira species identification. Infect Genet Evol 7:955–962.).
Many molecular DNA techniques have been applied to identify and classify the species and serovars of Leptospira (Ahmed et al., 2012Ahmed A, Grobusch MP, Klatser PR et al. (2012) Molecular Approaches in the Detection and Characterization of Leptospira. J Bacteriol Parasitol 3:2.). These include restriction endonuclease analysis (REA) of chromosomal DNA (Marshall et al., 1981Marshall RB, Wilton BE, Robinson AJ (1981) Identification of Leptospira serovars by restriction-endonuclease analysis. J Med Microbiol 14:163–166.), random amplified polymorphic DNA (RAPD) fingerprinting (Ramadass et al., 1997Ramadass P, Meerarani S, Venkatesha MD et al. (1997) Characterization of Leptospiral Serovars by Randomly Amplified Polymorphic DNA Fingerprinting. Int J Syst Bacteriol 47:575–576.), DNA-DNA hybridization (Yasuda et al., 1987Yasuda PH, Steigerwalt AG, Sulzer KR et al. (1987) Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol 37:407–415.; Brenner et al., 1999Brenner DJ, Kaufmann AF, Sulzer KR et al. (1999) Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol 49 Pt2:839–858.), arbitrarily primed PCR (Ramadass et al., 2002Ramadass P, Latha D, Senthilkumar A et al. (2002) Arbitrarily primed PCR- a rapid and simple method for typing of leptospiral serovars. Indian J Med Microbiol 20:25–28.), pulsed-field gel electrophoresis (PFGE) (Galloway and Levett, 2008Galloway RL, Levett PN (2008) Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars. Am J Trop Med Hyg 78:628–632.) and polymerase chain reaction (PCR) of specific genes followed by restriction fragment length polymorphism analysis (RFLP) (Li et al., 2009Li W, Raoult D, Fournier PE (2009) Bacterial strain typing in the genomic era. FEMS Microbiol Rev 33:892–916.). Recently, multilocus sequence typing has been applied as an alternative to immunological methods for the identification and classification of pathogenic leptospires (Ahmed et al., 2006Ahmed N, Devi SM, Valverde MA et al. (2006) Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob 5:28.; Pavan et al., 2008Pavan ME, Cairo F, Brihuega B et al. (2008) Multiple-locus variable-number tandem repeat analysis MLVA of Leptospira interrogans serovar Pomona from Argentina reveals four new genotypes. Comp Immunol Microbiol Infec Dis 31:37–45.; Leon et al., 2010Leon A, Pronost S, Fortier G et al. (2010) Multilocus Sequence Analysis for Typing Leptospira interrogans and Leptospira kirschneri. J Clin Microbiol 48:581–585.; Ahmed et al., 2011Ahmed A, Thaipadungpanit J, Boonsilp S et al. (2011) Comparison of two multilocus sequence based genotyping schemes for Leptospira species. PLoS Negl Trop Dis 5:e1374.; Boonsilp et al., 2013Boonsilp S, Thaipadungpanit J, Amornchai P et al. (2013) A Single Multilocus Sequence Typing (MLST) Scheme for Seven Pathogenic Leptospira Species. PLoS Negl Trop Dis 9:e824.). All of these techniques mentioned above have contributed significantly to the current taxonomic classification of the Leptospira genus (Morey et al., 2006Morey RE, Galloway RL, Bragg SL et al. (2006) Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44:3510–3516.; Slack et al., 2009Slack AT, Khairani-Bejo S, Symonds ML et al. (2009) Leptospira kmetyi sp. nov., isolated from an environmental source in Malaysia. Int J Syst Evol Microbiol 59:705–708.).
Quantitative DNA-DNA hybridization to measure genetic homology has been used as a reference for the classification of serovars within species (Yasuda et al., 1987Yasuda PH, Steigerwalt AG, Sulzer KR et al. (1987) Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol 37:407–415.; Perolat et al., 1998Perolat P, Chappel RJ, Adler B et al. (1998) Leptospira fainei sp. nov., isolated from pigs in Australia. Int J Syst Bacteriol 48:851–858., Brenner et al., 1999Brenner DJ, Kaufmann AF, Sulzer KR et al. (1999) Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol 49 Pt2:839–858.). However, this hybridization method is not routinely used for the identification of Leptospira species due its complex and laborious execution, which requires the use of radioactive isotopes and is therefore restricted to reference laboratories (Morey et al., 2006Morey RE, Galloway RL, Bragg SL et al. (2006) Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44:3510–3516.). It has also been observed that some serotypes are more characteristic of a single species, while others contain both pathogenic and nonpathogenic serovars (Morey et al., 2006Morey RE, Galloway RL, Bragg SL et al. (2006) Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44:3510–3516.). Furthermore, little correlation has been shown between serological classification and genotypic systems because a given serogroup can often be found in several species of Leptospira (Ahmed et al., 2012Ahmed A, Grobusch MP, Klatser PR et al. (2012) Molecular Approaches in the Detection and Characterization of Leptospira. J Bacteriol Parasitol 3:2.).
In addition to DNA-DNA hybridization and the other molecular methods mentioned above, specific PCR amplification of the 16S rRNA gene has contributed to the molecular characterization of some species of Leptospira (Ahmed et al., 2012Ahmed A, Grobusch MP, Klatser PR et al. (2012) Molecular Approaches in the Detection and Characterization of Leptospira. J Bacteriol Parasitol 3:2.). The advantage of this technique is that the use of a DNA template, particularly one designed based on the region that encodes the bacterially conserved 16S rRNA gene, can clearly reveal phylogenetic relationships among species (Morey et al., 2006Morey RE, Galloway RL, Bragg SL et al. (2006) Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44:3510–3516.).
La Scola et al. (2006a)La Scola B, Bui LTM, Baranton G et al. (2006a) Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263:142–147. have designed a universal primer pair for the identification of Leptospira species based on the gene encoding the β subunit of RNA polymerase (rpoB). These primers have been used to amplify and sequence the partial rpoB gene from 16 Leptospira species. According to the authors, analysis of the rpoB gene "may be useful as an initial screening test for the serovar identification of a new isolate of Leptospira and the detection or identification of Leptospira in clinical or environmental samples".
In previous studies, the utility of the rpoB gene for spirochete distinction among various bacterial species has been demonstrated (Renesto et al., 2000Renesto P, Lorvellec-Guillon K, Drancourt M et al. (2000) rpoB gene analysis as a novel strategy for identification of spirochetes from the genera Borrelia, Treponema and Leptospira. J Clin Microbiol 38:2200–2203.; Lee et al., 2000Lee SH, Kim BJ, Kim JH et al. (2000) Differentiation of Borrelia burgdorferi sensu lato on the basis of RNA polymerase gene (rpoB) sequences. J Clin Microbiol 38:2557–2562.; Khamis et al., 2004Khamis A, Raoult D, La Scola B (2004) rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol 42:3925–3931.; Balamurugan et al., 2013Balamurugan V, Gangadhar NL, Mohandoss N et al. (2013) Characterization of Leptospira isolates from animals and humans: phylogenetic analysis identifies the prevalence of intermediate species in India. Springer Plus 2:362.). Thus, the aim of this study was to investigate whether the PCR-amplified fragment of rpoB in conjunction with RFLP would allow for the determination of Leptospira serovars.
Material and Methods
Bacterial strains, media and growth conditions
For this study, sixty-eight Leptospira strains (Table 1) belonging to 11 reference species from the Pan American Institute for Food Protection and Zoonosis (INNPAZ) were used. Leptospires were grown for approximately five days at 30 °C in Ellinghausen-McCullough-Johnson-Harris (EMJH) culture medium (Difco) (Ellinghausen, 1973Ellinghausen HC (1973) Virulence, nutrition and antigenicity of Leptospira interrogans serotype Pomona in supplemented and nutrient deleted bovine albumin medium. Ann Microbiol (Paris) 124:477–493.).
Isolation of DNA
An one-mL aliquot of each Leptospira serovar was cultured in 5 mL EMJH medium for 7 to 10 days at 30 °C. The culture was then centrifuged at 3000 × g for 30 min, and DNA was extracted from the bacterial pellet by adding 1 mL lysozyme solution (10 mg/mL in TE buffer (10 mM Tris and 1 mM EDTA, pH 8.0) and Wizard Genomic DNA Purification System reagents according to the manufacturer's instructions (Promega Co.).
PCR assays
PCR amplification of a 600-bp region of the rpoB gene was performed with the primers 1900F (5′-CCTCATGGGTTCCAACATGCA-3′) and 2500R (5′-CGCATCCTCRAAGTTGTATTWCC-3′) as described by La Scola et al. (2006a)La Scola B, Bui LTM, Baranton G et al. (2006a) Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263:142–147.. Each PCR reaction contained 1.5 mM MgCl2, 200 μM dNTPs, 25–50 ng of DNA template, 1.5 units of Taq DNA polymerase, and 50 pmol of each primer. The PCR amplification reactions were carried out in a Veriti 96-well Thermal Cycler (Applied Biosystems) under the following conditions: an initial denaturation step of 2 min at 95 °C, 33 cycles of denaturation for 30 s at 94 °C, annealing at 51 °C for 30 s and extension at 72 °C for 2 min, with a final primer extension step for 10 min at 72 °C.
Restriction fragment length polymorphism (RFLP) analysis
To select enzymes for RFLP analysis, the results from in silico restriction digestion of twenty five rpoB sequences in GenBank® were analyzed with Webcutter 2.0 program (http://bio.lundberg.gu.se/cutter2/) to distinguish the generated fragments following separation by 6% polyacrylamide gel electrophoresis. The genomic sequences used were as follows: AE016823.1, L. interrogans serovar Copenhageni str. Fiocruz L1-130; AE010300.2, L. interrogans serovar Lai str. 56601; CP000350.1, L. borgpetersenii serovar Hardjo-bovis strain JB197; and CP000777.1, and L. biflexa serovar Patoc strain Patoc 1 (Ames). DNA sequences of the rpoB gene reported by La Scola et al. (2006a)La Scola B, Bui LTM, Baranton G et al. (2006a) Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263:142–147. and sequences obtained by us in this study were also used. These sequences were deposited in GenBank® under the accession numbers EU747300.1, EU747301.1, EU747302.1, EU747303.1, EU747304.1, EU747305.1, EU747306.1, EU747307.1, EU747308.1, EU747309.1, EU747310.1, EU747311.1, EU747312.1, EU747313.1, EU747314.1, EU747317.1, and EU747299.1, corresponding to L. interrogans serovar Bratislava, L. kirschneri serovar Grippotyphosa, L. borgpetersenii serovar Ballum, L. interrogans serovar Hardjo-prajitno, L. interrogans serovar Hebdomadis, L. borgpetersenii serovar Hardjo-bovis, L. interrogans serovar Pomona, L. borgpetersenii serovar Tarassovi, L. interrogans serovar Wolffi, L. biflexa serovar Andamana, L. borgpetersenii serovar Castellonis, L. borgpetersenii serovar Sejroe, L. interrogans serovar Djasiman, L. interrogans serovar Schueffneri, L. borgpetersenii serovar Whitticombi, L. interrogans serovar Sentot, and L. interrogans serovar Canicola, respectively.
PCR products were subjected to restriction digestion with TaqI, Tru1I, Sau3AI and MslI endonucleases (Promega Co.) for 3 h at the recommended temperatures. To calculate the relative molecular masses of the digested fragments, a 100-bp DNA Ladder was used (Promega Co.). The digestion and separation of the DNA fragments by 6% polyacrylamide gel electrophoresis were repeated at least three times for all serovars to establish the final restriction patterns.
Dendrogram construction
LabImage version 2.7.0 software (Kapelan GMBH) was used for constructing a binary matrix scored on the presence (1) or absence (2) of each fragment generated by PCR-RFLP with the rpoB primers. Cluster analysis based on similarity (Nei, 1972Nei M (1972) Genetic distance between populations. Am Nat 106:283–292.) was performed by the unweighted pair group method (UPGMA) with the arithmetic averages clustering algorithm (Sneath and Sokal, 1973Sneath PHA, Sokal RR (1973) Numerical Taxonomy. Freeman, San Francisco, C.A.), and the randomization procedure implemented in Tools for Population Genetic Analysis (TFPGA) software package according to Miller (1998)Miller MP (1998) TFPGA: Tools for Population Genetic Analyses for Windows. Arizona State University, USA. was used to construct the dendrogram.
Results
In silico analysis of rpoB sequences deposited in GenBank indicated that a combination of four possible restriction enzymes was necessary to distinguish the Leptospira serovars as follows: TaqI, Tru1I, Sau3AI and MslI. Alone, each enzyme was able to identify only one or two different serovars.
Digestion with TaqI resulted in ten different patterns (A to J), which are schematically represented in Table 2 and had the following frequencies: A, 29.4% (20); B, 10.3% (7); C, 7.35% (5); D, 13.2% (9); E, 11.8% (8); F, 4.41% (3); G, 16.2% (11); H, 7.35% (3); I, 1.47% (1); and J, 1.47% (1). Thus, TaqI identified two serovars, Huallaga of L. noguchii (profile I) and Alice of L. santarosai (profile J), as shown in Figure 1. The G profile pattern was observed in almost all L. santarosai serovars, with the exception of the serovar Alexi (profile D), and it was only identified in the Muenchen serovar L. interrogans.
Restriction patterns of the 600-bp fragment of the rpoB gene of Leptospira following digestion with TaqI, Tru1I, Sau3AI, and MslI endonucleases.
Polyacrylamide gel electrophoresis (6%) of the PCR products resulting from the digestion of the rpoB gene with the restriction endonuclease TaqI. was consistent with the 100-bp molecular weight ladder.
The Tru1I enzyme also exhibited ten distinct restriction patterns (A to J) with the following frequencies: A, 2.94% (2); B, 23.5% (16); C, 16.2% (11); D, 11.8% (8); E, 22.1% (15); F, 14.7% (10); G, 2.94% (2); H, 2.94% (2); I, 1.47% (1); and J, 1.47% (1). These patterns are summarized in Table 2 and identified the serovars Huallaga of L. noguchii (profile I) and Trinidad of L. santarosai (profile J).
The combination of both enzymes, TaqI and Tru1I, generated 23 distinct patterns with some interesting results as follows: profile A of TaqI and Tru1I (profile AA) was species-specific and was only observed for L. biflexa. Profile AC was displayed by all serovars of L. kirschneri and by serovar Hualien of L. terpstrae; therefore, it is nearly species-specific. Finally, the profiles AG, FG, FE, CE, FD, BE and GE were unique to the serovars Mini, Kaup, Lanka, Szwajizak, Waskurin, Myocastoris and Maru, respectively.
Digestion with the Sau3AI enzyme generated nine distinct restriction patterns, which are summarized in Table 2 with the following frequencies: A, 2.94% (2); B, 22.1% (15); C, 30.9% (21); D, 4.41% (3); E, 7.35% (5); F, 16.2% (11); G, 8.82% (6); H, 5.88% (4); and I, 1.47% (1). Sau3AI digestion identified only the serovar Ranarum to have a serovar-specific profile. However, the combination of all three enzymes generated 30 distinct profiles, including EEE, DFD, ACC, HDD, GBB and AHH, which were specific for the serovars Whitcomb, Icterohaemorrhagiae, Hardjo, Ramisi, Semaranga, Vargonicas and Sarmin, respectively.
Finally, digestion with the enzyme MslI produced only five distinct restriction patterns, which are summarized in Table 2 with the following frequencies: A, 10.3% (7); B, 20.6% (14); C, 57.4% (39); D, 10.3% (7); and E, 1.47% (1). Only the serovar Saopaulo was identified by this enzyme to have a serovar-specific profile.
The combination of the four enzymes TaqI, Tru1I, Sau3AI and MslI generated 35 distinct profiles and identified the serovars Parameles (EFFD) and Celledoni (HDFA). In addition, this combination helped to distinguish the serovars Valbuzzi and Tropica, which had the profiles DFFC and GBCC, respectively (Table 3).
Grouping of the serovars, serogroups and species of the Leptospira genus based on the restriction patterns generated with the four endonucleases.
Out of sixty-eight serovars analyzed for RFLP polymorphisms in the region of the coding sequence containing the β-subunit gene of RNA polymerase, 22 serovars from nine species (32%) were identified by digestion with the enzymes TaqI, Tru1I, Sau3AI and MslI (Table 3), and the other 46 strains were clustered into 13 groups with two to seven serovars.
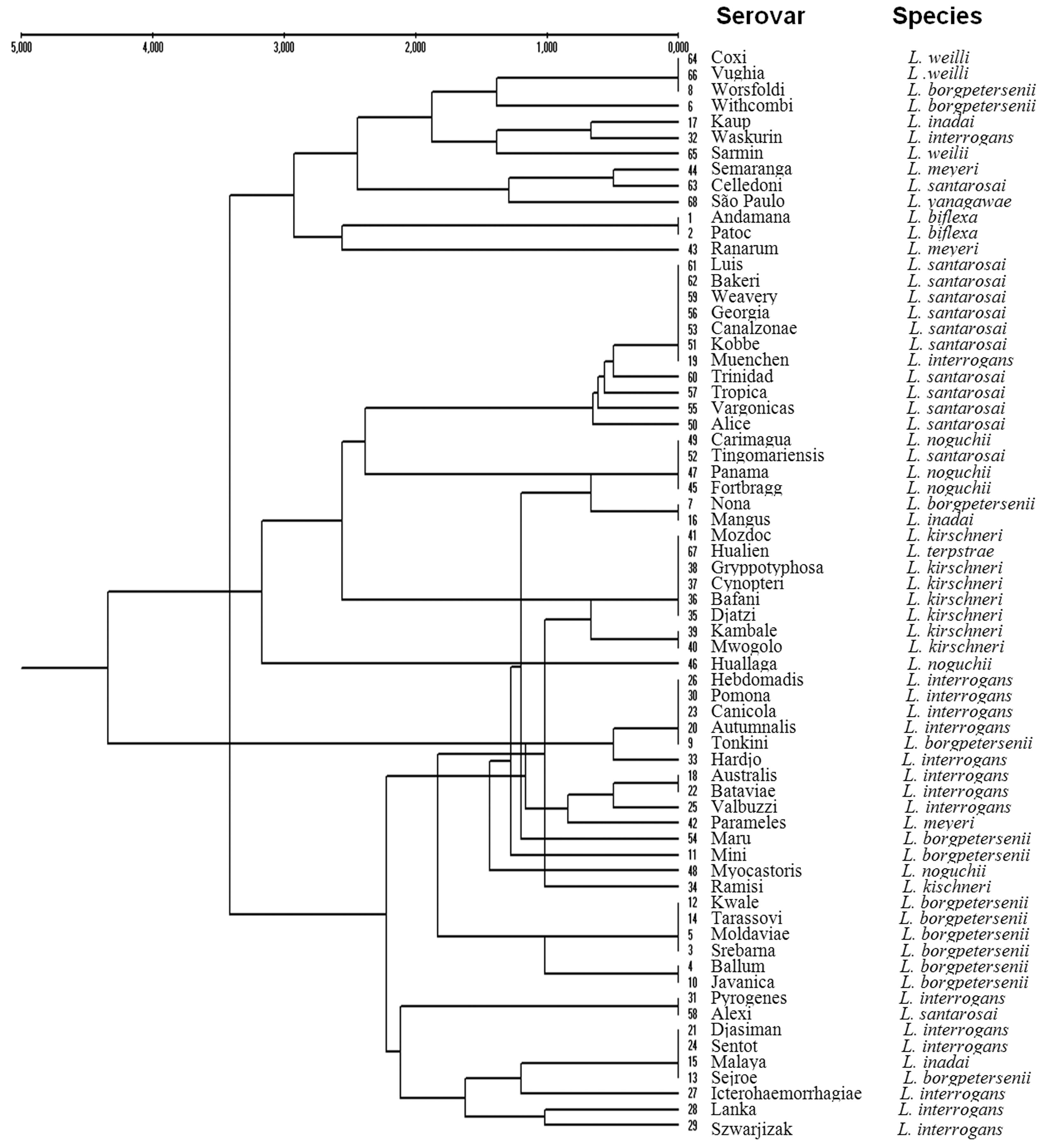
A dendrogram obtained from a matrix constructed with the results from the fragments generated by PCR-RFLP with the four restriction endonucleases (Figure 2) showed clustering of the sixty-eight reference serovars. Several of the tested strains appeared to be distant from others of the same species in relation to the current taxonomic classification. The serovar Kaup (L. inadai) was grouped with the serovar Waskurin (L. interrogans); the serovar Ranarum (L. meyeri) was similar to the nonpathogenic L. biflexa; the serovar Muenchen (L. interrogans) clustered with those of L. santarosai; the serovar Nona (L. borgpetersenii) was closer to the serovar Mangus (L. inadai); the serovar Hualien (L. terpstrae) grouped with the those of L. kirschneri; the Huallaga and Myocastoris serovars (L. noguchii) were located in different branches; the serovar Tonkini (L. borgpetersenii) grouped with the majority of those of L. interrogans; the serovar Ramisi (L. kirschneri) was closer to those of L. borgpetersenii; and the serovar Alexi (L. santarosai) was grouped with those of Djasiman, Pyrogenes and Sentot (L. interrogans), Malaya (L. inadai) and Sejroe (L. borgpetersenii).
Dendrogram constructed by joint analysis of the bands generated by the restriction endonucleases TaqI, Tru1I, Sau3AI, and MslI.
Discussion
The correlation between the serological and genotypic classifications of leptospires is low, and identification is complicated because the same serovar can be distributed among different species (Ahmed et al., 2012Ahmed A, Grobusch MP, Klatser PR et al. (2012) Molecular Approaches in the Detection and Characterization of Leptospira. J Bacteriol Parasitol 3:2.; Balamurugan et al., 2013Balamurugan V, Gangadhar NL, Mohandoss N et al. (2013) Characterization of Leptospira isolates from animals and humans: phylogenetic analysis identifies the prevalence of intermediate species in India. Springer Plus 2:362.). It is assumed that this lack of correlation between species and serovars is the result of horizontal transference between species of the genes that determine serotypes, but the basis of this transference, which is responsible for exchanging genetic determinants, is still unknown (Cerqueira and Picardeau, 2009Cerqueira G, Picardeau M (2009) A century of Leptospira strain typing. Infect Genet Evol 9:760–768.). A single base difference differentiated many strains of L. interrogans and L. kirschneri; therefore, phylogenetic representation may be less meaningful than sequence identities at variable positions (Cerqueira and Picardeau, 2009Cerqueira G, Picardeau M (2009) A century of Leptospira strain typing. Infect Genet Evol 9:760–768.).
The aim of this work was to identify Leptospira strains at the serovar level by performing PCR-RFLP to amplify a 600-bp fragment of the coding sequence of the β subunit of the RNA polymerase gene. The rpoB gene has been widely studied in other organisms and is considered by many researchers to be more useful than the 16S ribosomal RNA gene for the differentiation of bacterial species (La Scola et al., 2006aLa Scola B, Bui LTM, Baranton G et al. (2006a) Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263:142–147.; Ahmed et al., 2006Ahmed N, Devi SM, Valverde MA et al. (2006) Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob 5:28.; Macheras et al., 2011Macheras E, Roux AL, Bastian S et al. (2011) Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J Clin Microbiol 49:491–499.; Ahmed et al., 2012Ahmed A, Grobusch MP, Klatser PR et al. (2012) Molecular Approaches in the Detection and Characterization of Leptospira. J Bacteriol Parasitol 3:2.). In addition, twenty-five sequences of the rpoB gene of Leptospira are already available in databases, thereby facilitating access and minimizing project costs.
In a previous report, La Scola et al. (2006a)La Scola B, Bui LTM, Baranton G et al. (2006a) Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263:142–147. have compared similarities in the rrs and rpoB genes between different Leptospira serovars. Using the rpoB gene, they were able to effectively distinguish 11 of 19 serovars tested, differentiating them from other species and showing greater numbers of polymorphisms in both genes, leading to the conclusion that the rpoB gene could distinguish species with a higher number of differences between base pairs.
In this study, 68 Leptospira serovars were analyzed for polymorphisms in a specific region of the rpoB gene using the endonucleases TaqI, Tru1I, Sau3AI and MslI. These enzymes were selected after in silico restriction digestion of the rpoB sequences deposited in GenBank. We were able to identify 22 strains from nine species at the serovar level (32%). The rpoB gene has been widely used as an alternative tool in the phylogeny and identification of different species of bacteria, such as Coxiella burnetii (Mollet et al., 1998Mollet C, Drancourt M, Raoult D (1998) Determination of Coxiellaburnetii rpoB sequence and its use for phylogenetic analysis. Gene 207:97–103.), Afipia (Khamis et al., 2003Khamis A, Colson P, Raoult D et al. (2003) Usefulness of rpoB gene sequencing for identification of Afipia and Bosea species, including a strategy for choosing discriminative partial sequences. Appl Environ Microbiol 69:6740–6749.), Mycoplasma (Kim et al., 2003Kim KS, Ko KS, Chang MW et al. (2003) Use of rpoB sequences for phylogenetic study of Mycoplasma species. FEMS Microbiol Lett 226:299–305.), Corynebacterium (Khamis et al., 2004Khamis A, Raoult D, La Scola B (2004) rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol 42:3925–3931.), Acinetobacter (La Scola et al., 2006bLa Scola B, Gundi VA, Khamis A et al. (2006b) Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 44:827–832.), Mycobacterium (Adekambi et al., 2006Adekambi T, Berger P, Raoult D et al. (2006) rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp.nov. Int J Syst Evol Microbiol 56:133–143.; Ben et al., 2008Ben Salah I, Adekambi T, Raoult D et al. (2008) rpoB sequence-based identification of Mycobacterium avium complex species. Microbiol 154:3715–3723.), Halobacterium (Minegishi et al., 2010Minegishi H, Kamekura M, Itoh T et al. (2010) Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B9 (rpoB9) gene. Int J Syst Evol Microbiol 60:2398–2408.) and Cyanobacteria (Gaget et al., 2011Gaget V, Gribaldo S, Tandeau de Marsac N (2011) A rpoB signature sequence provides unique resolution for the molecular typing of cyanobacteria. Int J Syst Evol Microbiol 61:170–183.).
In a recent study, the rpoB gene has been successfully used to identify or detect Leptospira species in animals and humans in India (Balamurugan et al., 2013Balamurugan V, Gangadhar NL, Mohandoss N et al. (2013) Characterization of Leptospira isolates from animals and humans: phylogenetic analysis identifies the prevalence of intermediate species in India. Springer Plus 2:362.). Because each Leptospira serovar is associated with specific host symptoms, their identification is essential for the development of epidemiological studies (Cerqueira and Picardeau, 2009Cerqueira G, Picardeau M (2009) A century of Leptospira strain typing. Infect Genet Evol 9:760–768., Li et al., 2009Li W, Raoult D, Fournier PE (2009) Bacterial strain typing in the genomic era. FEMS Microbiol Rev 33:892–916.).
Clustering analysis of the results of this study correctly grouped 22 serovars by species. Considerable similarities in the analyzed genomic region were observed among all serovars. Analysis of dendrograms constructed from the results of each restriction enzyme and from the collective results for all of the enzymes showed the formation of clusters, for which serovars of various species had identical profiles. The groups formed by the rpoB gene profiles showed varying degrees of similarity and clade formation. Based on this, similar banding patterns were observed among the serovars Mangus, Nona, Alexi, Pyrogenes, Sentot, Malaya and Sejroe, despite the fact that they belonged to different species. These findings are in accordance with similar dendrogram analyses reported previously (Perolat et al., 1998Perolat P, Chappel RJ, Adler B et al. (1998) Leptospira fainei sp. nov., isolated from pigs in Australia. Int J Syst Bacteriol 48:851–858.; Morey et al., 2006Morey RE, Galloway RL, Bragg SL et al. (2006) Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44:3510–3516.; Cerqueira and Picardeau, 2009Cerqueira G, Picardeau M (2009) A century of Leptospira strain typing. Infect Genet Evol 9:760–768.; Balamurugan et al., 2013Balamurugan V, Gangadhar NL, Mohandoss N et al. (2013) Characterization of Leptospira isolates from animals and humans: phylogenetic analysis identifies the prevalence of intermediate species in India. Springer Plus 2:362.), showing similar cluster formations and variations in serovar-species grouping.
The addition of new enzymes for the production of additional profiles should clarify the positions of other serovars. Still, these results suggest that the use of this technique to assess gene sequences may reveal a precise sensu stricto classification of these serovars.
Molecular techniques have been used for the characterization of Leptospira isolates; however, most can only make identifications to the species level (Galloway and Levett, 2010Galloway RL, Levett PN (2010) Application and Validation of PFGE for Serovar Identification of Leptospira Clinical Isolates. PLoS Negl Trop Dis 14:e824.), such as 16S rRNA sequence analysis (Morey et al., 2006Morey RE, Galloway RL, Bragg SL et al. (2006) Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44:3510–3516.), RFLP (Li et al., 2009Li W, Raoult D, Fournier PE (2009) Bacterial strain typing in the genomic era. FEMS Microbiol Rev 33:892–916.) and MLST (Boonsilp et al., 2013Boonsilp S, Thaipadungpanit J, Amornchai P et al. (2013) A Single Multilocus Sequence Typing (MLST) Scheme for Seven Pathogenic Leptospira Species. PLoS Negl Trop Dis 9:e824.). PFGE has demonstrated the reliable and reproducible identification of Leptospira at the serovar level (Galloway and Levett, 2010Galloway RL, Levett PN (2010) Application and Validation of PFGE for Serovar Identification of Leptospira Clinical Isolates. PLoS Negl Trop Dis 14:e824.). These approaches have greatly contributed to a revolution in both Leptospira detection and characterization (Ahmed et al., 2012Ahmed A, Grobusch MP, Klatser PR et al. (2012) Molecular Approaches in the Detection and Characterization of Leptospira. J Bacteriol Parasitol 3:2.). On the other hand, the molecular tools described so far for the characterization of Leptospira suffer from significant drawbacks. For example, PFGE, RFLP, and REA require large quantities of purified DNA, have low levels of discrimination, produce data that is difficult to interpret, suffer from a lack of reproducibility and require specialized equipment (Ahmed et al., 2006Ahmed N, Devi SM, Valverde MA et al. (2006) Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob 5:28.).
Notably, the 16S rRNA gene has been considered the gold standard in molecular surveys of bacterial and archaeal diversity, but it has several disadvantages as follows: it is often present in multiple copies, has little resolution below the species level and cannot be readily interpreted in an evolutionary framework (Vos et al., 2012Vos M, Quince C, Pijl AS et al. (2012) A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity. PLoS One 2:e30600.).
The main advantages of the use of the rpoB gene over the 16S rRNA gene are as follows: (i) it is universally present in all prokaryotes; (ii) it typically occurs in a single-copy, essential protein-encoding gene, and sequence errors can be readily identified and removed if they introduce disruptions in the reading frame; (iii) it possesses both slowly and quickly evolving regions, enabling the design of probes and primers of differing specificities; (iv) it has a housekeeping function, making it less susceptible to some forms of lateral gene transfer; and (v) it is large enough in size to contain phylogenetic information, even after the removal of regions that are difficult to align (Case et al., 2007Case RJ, Boucher Y, Dahllöf I et al. (2007) Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol 1:278–288.; Vos et al., 2012Vos M, Quince C, Pijl AS et al. (2012) A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity. PLoS One 2:e30600.).
Our findings "in vitro" indicate that the PCR-RFLP technique is a powerful and reproducible test that may be used as a complement or alternative tool to assess the distribution of Leptospira strains within species. Additionally, we recommend the use of PCR-RFLP with in silico digestion of the polymorphic sequences of other conserved genes already deposited in GenBank as a promising technique for the genomic classification of the Leptospira genus.
Conclusion
This study demonstrated that PCR-RFLP is practical and efficient, enabling the differentiation of species and serovars with good discriminatory power, reproducibility and easily interpretable results. In addition, this method is cost-effective for most research laboratories. This technique has also been shown to be suitable for phylogenetic studies and the classifications of species, serovars and strains. The selected 600-bp polymorphic sequence of the rpoB gene produced restriction profiles that allowed for the accurate and timely identification of 32% of the 68 tested strains. We demonstrated that this approach achieves the stated purpose and that serological typing is unreliable for the classification of pathogenic Leptospira. However, additional studies should be undertaken to reclassify these serovars within the species with which they have greater genotypic affinities based on analysis of hypervariable regions of multiple housekeeping genes and especially to investigate whether the clinical leptospirosis symptoms induced by these serovars are presented according to the species with which they are most phylogenetically related.
Acknowledgments
This study was supported by the Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Fundação de Amparo à Pesquisa e Desenvolvimento Científico do Maranhão (FAPEMA) and the PRPq/UFMG. We thank Dr. Élvio C. Moreira from the Department of Preventive Medicine of the Veterinary School of UFMG for providing the Leptospira strains and Dr. Regina M. Nardi Drummond and Dr. Vera Lúcia dos Santos for their assistance at the laboratory facilities with the growing and maintenance of the Leptospira strains.
References
- Adekambi T, Berger P, Raoult D et al. (2006) rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp.nov. Int J Syst Evol Microbiol 56:133–143.
- Ahmed N, Devi SM, Valverde MA et al. (2006) Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob 5:28.
- Ahmed A, Anthony RM, Hartskeerl RA (2010) A simple and rapid molecular method for Leptospira species identification. Infect Genet Evol 7:955–962.
- Ahmed A, Thaipadungpanit J, Boonsilp S et al. (2011) Comparison of two multilocus sequence based genotyping schemes for Leptospira species. PLoS Negl Trop Dis 5:e1374.
- Ahmed A, Grobusch MP, Klatser PR et al. (2012) Molecular Approaches in the Detection and Characterization of Leptospira J Bacteriol Parasitol 3:2.
- Balamurugan V, Gangadhar NL, Mohandoss N et al. (2013) Characterization of Leptospira isolates from animals and humans: phylogenetic analysis identifies the prevalence of intermediate species in India. Springer Plus 2:362.
- Ben Salah I, Adekambi T, Raoult D et al. (2008) rpoB sequence-based identification of Mycobacterium avium complex species. Microbiol 154:3715–3723.
- Bharti AR, Nally JE (2003) Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3:757–771.
- Brenner DJ, Kaufmann AF, Sulzer KR et al. (1999) Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol 49 Pt2:839–858.
- Boonsilp S, Thaipadungpanit J, Amornchai P et al. (2013) A Single Multilocus Sequence Typing (MLST) Scheme for Seven Pathogenic Leptospira Species. PLoS Negl Trop Dis 9:e824.
- Case RJ, Boucher Y, Dahllöf I et al. (2007) Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol 1:278–288.
- Cerqueira G, Picardeau M (2009) A century of Leptospira strain typing. Infect Genet Evol 9:760–768.
- Ellinghausen HC (1973) Virulence, nutrition and antigenicity of Leptospira interrogans serotype Pomona in supplemented and nutrient deleted bovine albumin medium. Ann Microbiol (Paris) 124:477–493.
- Gaget V, Gribaldo S, Tandeau de Marsac N (2011) A rpoB signature sequence provides unique resolution for the molecular typing of cyanobacteria Int J Syst Evol Microbiol 61:170–183.
- Galloway RL, Levett PN (2008) Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars. Am J Trop Med Hyg 78:628–632.
- Galloway RL, Levett PN (2010) Application and Validation of PFGE for Serovar Identification of Leptospira Clinical Isolates. PLoS Negl Trop Dis 14:e824.
- Khamis A, Colson P, Raoult D et al. (2003) Usefulness of rpoB gene sequencing for identification of Afipia and Bosea species, including a strategy for choosing discriminative partial sequences. Appl Environ Microbiol 69:6740–6749.
- Khamis A, Raoult D, La Scola B (2004) rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol 42:3925–3931.
- Kim KS, Ko KS, Chang MW et al. (2003) Use of rpoB sequences for phylogenetic study of Mycoplasma species. FEMS Microbiol Lett 226:299–305.
- Ko AI, Goarant C, Picardeau M (2009) Leptospira: The Dawn of the Molecular Genetics Era for an Emerging Zoonotic Pathogen. Nat Rev Microbiol 10:736–747.
- La Scola B, Bui LTM, Baranton G et al. (2006a) Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263:142–147.
- La Scola B, Gundi VA, Khamis A et al. (2006b) Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 44:827–832.
- Lau C, Clements A, Skelly C et al. (2012) Leptospirosis in American Samoa-Estimating and mapping risk using environmental data. PLoS Negl Trop Dis 6:e1669.
- Lee SH, Kim BJ, Kim JH et al. (2000) Differentiation of Borrelia burgdorferi sensu lato on the basis of RNA polymerase gene (rpoB) sequences. J Clin Microbiol 38:2557–2562.
- Leon A, Pronost S, Fortier G et al. (2010) Multilocus Sequence Analysis for Typing Leptospira interrogans and Leptospira kirschneri J Clin Microbiol 48:581–585.
- Li W, Raoult D, Fournier PE (2009) Bacterial strain typing in the genomic era. FEMS Microbiol Rev 33:892–916.
- Macheras E, Roux AL, Bastian S et al. (2011) Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J Clin Microbiol 49:491–499.
- Marshall RB, Wilton BE, Robinson AJ (1981) Identification of Leptospira serovars by restriction-endonuclease analysis. J Med Microbiol 14:163–166.
- Miller MP (1998) TFPGA: Tools for Population Genetic Analyses for Windows. Arizona State University, USA.
- Minegishi H, Kamekura M, Itoh T et al. (2010) Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B9 (rpoB9) gene. Int J Syst Evol Microbiol 60:2398–2408.
- Mohammed H, Nozha C, Hakim K et al. (2011) Leptospira: Morphology, Classification and Pathogenesis. Bacteriol Parasitol 2:6.
- Mollet C, Drancourt M, Raoult D (1998) Determination of Coxiellaburnetii rpoB sequence and its use for phylogenetic analysis. Gene 207:97–103.
- Morey RE, Galloway RL, Bragg SL et al. (2006) Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44:3510–3516.
- Nei M (1972) Genetic distance between populations. Am Nat 106:283–292.
- Pavan ME, Cairo F, Brihuega B et al. (2008) Multiple-locus variable-number tandem repeat analysis MLVA of Leptospira interrogans serovar Pomona from Argentina reveals four new genotypes. Comp Immunol Microbiol Infec Dis 31:37–45.
- Perolat P, Chappel RJ, Adler B et al. (1998) Leptospira fainei sp. nov., isolated from pigs in Australia. Int J Syst Bacteriol 48:851–858.
- Ramadass P, Meerarani S, Venkatesha MD et al. (1997) Characterization of Leptospiral Serovars by Randomly Amplified Polymorphic DNA Fingerprinting. Int J Syst Bacteriol 47:575–576.
- Ramadass P, Latha D, Senthilkumar A et al. (2002) Arbitrarily primed PCR- a rapid and simple method for typing of leptospiral serovars. Indian J Med Microbiol 20:25–28.
- Renesto P, Lorvellec-Guillon K, Drancourt M et al. (2000) rpoB gene analysis as a novel strategy for identification of spirochetes from the genera Borrelia, Treponema and Leptospira J Clin Microbiol 38:2200–2203.
- Slack AT, Khairani-Bejo S, Symonds ML et al. (2009) Leptospira kmetyi sp. nov., isolated from an environmental source in Malaysia. Int J Syst Evol Microbiol 59:705–708.
- Sneath PHA, Sokal RR (1973) Numerical Taxonomy. Freeman, San Francisco, C.A.
- Vos M, Quince C, Pijl AS et al. (2012) A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity. PLoS One 2:e30600.
- World Health Organization. Leptospirosis worldwide. (1999). Weekly Epidemiol Rec 74:237–242.
- World Health Organization (2006) Informal Consultation on Global Burden of Leptospirosis: Methods of Assessment. Available at: http://www.who.int/entity/foodsafety/zoonoses/InformalConsultationOnBoDLeptospirosis.pdf
» http://www.who.int/entity/foodsafety/zoonoses/InformalConsultationOnBoDLeptospirosis.pdf - Yasuda PH, Steigerwalt AG, Sulzer KR et al. (1987) Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol 37:407–415.
Publication Dates
-
Publication in this collection
Apr-Jun 2015
History
-
Received
03 Sept 2012 -
Accepted
29 Oct 2014