Abstract
Multiple-locus variable number-tandem repeat analysis (MLVA) of Mycobacterium avium subspecies paratuberculosis (MAP) isolates may contribute to the knowledge of strain diversity in Argentina. Although the diversity of MAP has been previously investigated in Argentina using IS900-RFLP, a small number of isolates were employed, and a low discriminative power was reached. The aim of the present study was to test the genetic diversity among MAP isolates using an MLVA approach based on 8 repetitive loci. We studied 97 isolates from cattle, goat and sheep and could describe 7 different patterns: INMV1, INMV2, INMV11, INMV13, INMV16, INMV33 and one incomplete pattern. INMV1 and INMV2 were the most frequent patterns, grouping 76.3% of the isolates. We were also able to demonstrate the coexistence of genotypes in herds and co-infection at the organism level. This study shows that all the patterns described are common to those described in Europe, suggesting an epidemiological link between the continents.
paratuberculosis; molecular typing; MIRU-VNTR; MLVA
Introduction
Johne's disease or paratuberculosis (PTB) is an enteric disease aetiologically associated with Mycobacterium avium subsp. paratuberculosis (MAP). This chronic and progressive intestinal infection causes important economic losses to dairy and beef herds around the world (OIE, 2009OIE. 2009. World Animal Health Information Database (WAHID) - Version: 1.4 Available at http://www.oie.int.
http://www.oie.int...
). Moreover, its zoonotic potential, an issue currently under discussion, is also considered a major concern. MAP has been largely described as restricted to ruminants, such as goats, sheep, cattle, deer, and bison, and recently in camelids (Buergelt et al., 2000Buergelt CD, Layton AW, Ginn PE et al. (2000) The pathology of spontaneous paratuberculosis in the North American bison (Bison bison). Vet Pathol 37:428–438.; Djønne et al., 2005Djønne B, Pavlik I, Svastova P et al. (2005) IS900 Restriction Fragment Length Polymorphism (RFLP) Analysis of Mycobacterium avium subsp. paratuberculosis isolates from goats and cattle in Norway. Acta Vet Scand 46:13–18.; Salgado et al., 2009Salgado M, Herthnek D, Bölske G et al. (2009) First isolation of Mycobacterium avium subsp. paratuberculosis from wild guanacos (Lama guanicoe) on Tierra del Fuego Island. J Wildl Dis 45:295–301.; Attili et al., 2011Attili AR, NguNgwa V, Preziuso S et al. (2011) Ovine paratuberculosis: A seroprevalence study in dairy flocks reared in the Marche region, Italy. Vet Med Int doi:10.4061/2011/782875.
https://doi.org/10.4061/2011/782875...
; Ghosh et al., 2012Ghosh P, Hsu C, Alyamani EJ et al. (2012) Genome-wide analysis of the emerging infection with Mycobacterium avium subspecies paratuberculosis in the Arabian camels (Camelus dromedarius). PLoS One doi: 10.1371/journal.pone.0031947.
https://doi.org/10.1371/journal.pone.003...
). Despite this, the isolation of MAP from non-ruminant wildlife, in some cases with pathological lesions, may constitute supporting evidence of a widening of the host range and reservoirs (Beard et al., 2001Beard PM, Daniels MJ, Henderson D et al. (2001) Paratuberculosis infection of nonruminant wildlife in Scotland. J Clin Microbiol 39:1517–1521.; Stevenson et al., 2009Stevenson K, Alvarez J, Bakker D et al. (2009) Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol doi: 10.1186/1471-2180-9-212.
https://doi.org/10.1186/1471-2180-9-212...
). Their long-term survival in soil, grass, water samples and biofilms (Whittington et al., 2004Whittington RJ, Marshall DJ, Nicholls PJ et al. (2004) Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl Environ Microbiol 70:2989–3004.; Pickup et al., 2005Pickup RW, Rhodes G, Arnott S et al. (2005) Mycobacterium avium subsp. paratuberculosis in the catchment area and water of the river Taff in South Wales, United Kingdom, and its potential relationship to clustering of Crohn's disease cases in the city of Cardiff. Appl Environ Microbiol 71:2130–2139., Cook et al., 2010Cook KL, Britt JS, Bolster CH (2010) Survival of Mycobacterium avium subsp. paratuberculosis in biofilms on livestock watering trough materials. Vet Microbiol 141:103–109.), replication and persistence in amoeba (Mura et al., 2006Mura M, Bull TJ, Evans H et al. (2006) Replication and long-term persistence of bovine and human strains of Mycobacterium avium subsp. paratuberculosis within Acanthamoeba polyphaga. Appl Environ Microbiol 72:854–859.) constitute important evidence that the environment (natural or artificial) may be a major source of MAP strains. These factors increase the exposure of animals and humans to this pathogen and could contribute to the endemicity of PTB. Although epidemiological data are scarce in Argentina, PTB has been well characterised in Buenos Aires province, one of the most productive regions of the country, where a seroprevalence ranging from 7.2 to 19.6% in cattle herds has been reported in the Salado river basin region (INTA-Balcarce, data not published). Additionally, the disease has been widely described in different hosts such as sheep (Nader and Bernardelli, 1986Nader A, Bernardelli A (1986) Utilización del test tuberculínico intradérmico comparativo en ovinos como prueba de screening en paratuberculosis. Revista de Medicina Veterinaria 67:335–338.; Jorge et al., 2000Jorge MC, Schettino DM, Torres P et al. (2000) Primera descripción de infección concomitante de tuberculosis y paratuberculosis en ovinos lecheros en Argentina. Rev Sci Tech Off Int Epizoot 19:800–809.; Bernardelli et al., 2002Bernardelli A, Cicuta NE, Nicola A et al. (2002) Ovine paratuberculosis: field diagnosis. 7th International Colloquium on Paratuberculosis Bilbao, Spain.), cattle (Moreira et al., 1999Moreira AR, Paolicchi F, Morsella C et al. (1999) Distribution of IS900 restriction fragment length polymorphism types among animal Mycobacterium avium subsp. paratuberculosis isolates from Argentina and Europe. Vet Microbiol 70: 251–259.; Paolicchi et al., 2003Paolicchi F, Zumárraga J, Gioffré A et al. (2003) Application of different methods for the diagnosis of paratuberculosis in a dairy cattle herd in Argentina. J Vet Med 50:20–26.), captive deer (Paolicchi et al., 2001Paolicchi FA, Vagnozzi A, Morsella CG et al. (2001) Paratuberculosis in red deer (Cervus elaphus): an immunohistochemical study. J Vet Med, Series B 48:313–320.; Verna et al., 2002Verna A, Morsella C, Zumárraga M et al. (2002) Strains characterization by PCR and RFLP from Mycobacterium avium subsp. paratuberculosis isolates of red deer with paratuberculosis. 7th International Colloquium on Paratuberculosis, Bilbao, Spain.), a captive mouflon (Ovis musimon) (Bernardelli et al., 2005Bernardelli A, Zumárraga M, Alonso B et al. (2005) Paratuberculosis in Mouflon (Ovis musimon). 8th International Colloquium on Paratuberculosis, Copenhagen, Denmark.) and goats (Fiorentino et al., 2012Fiorentino MA, Gioffré A, Cirone K et al. (2012) First isolation of Mycobacterium avium subsp. paratuberculosis in a dairy goat in Argentina: Pathology and molecular characterization. Small Ruminant Res http://dx.doi.org/10.1016/j.smallrumres.2012.06.010.
http://dx.doi.org/10.1016/j.smallrumres....
). Several factors such as limitations in diagnostic availability all over the country, the absence of an official infection control programme and the intensification of livestock production systems lead to speculation that PTB will be more widespread in the country in the future. In contrast, bovine tuberculosis (bTB) is better characterised at the epidemiological level in our country. However, although the voluntary official control programme has been successfully applied since 1999, the region is not considered bTB-free. As a consequence, co-infection with M. bovis and MAP is plausible in Argentinean herds and flocks (Jorge et al., 2000Jorge MC, Schettino DM, Torres P et al. (2000) Primera descripción de infección concomitante de tuberculosis y paratuberculosis en ovinos lecheros en Argentina. Rev Sci Tech Off Int Epizoot 19:800–809.). Furthermore, PTB also appears to be a possible interference in bTB diagnosis, which has not been estimated in our country. A previous study in the late 1990s performed in bovine and farmed deer isolates from our country suggested a low epidemiological association with Europe. In that study, Moreira and co-workers typed 61 MAP isolates from cattle and deer by IS900-restriction fragment polymorphisms analysis (RFLP), and four main patterns were found. The pattern named A was identical to a less frequent pattern, R9 (C17), from Europe, whereas the rest were not found on the same continent (Moreira et al., 1999Moreira AR, Paolicchi F, Morsella C et al. (1999) Distribution of IS900 restriction fragment length polymorphism types among animal Mycobacterium avium subsp. paratuberculosis isolates from Argentina and Europe. Vet Microbiol 70: 251–259.). Molecular typing based on MIRU (Mycobacterial interspersed repetitive unit) and VNTR (variable number in tandem repeat) loci has been employed as a simple and rapid procedure for differentiation between MAP and other M. avium complex isolates (Bull et al., 2003Bull TJ, Sidi-Boumedine K, McMinn EJ et al. (2003) Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol Cell Probes 17:157–164.; Radomski et al., 2010Radomski N, Thibault VC, Karoui C et al. (2010) Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol 48:1026–1034.). PCR-based genotyping is rapid and inexpensive, and its discriminative power is similar to that previously reported for other techniques such as RFLP (Motiwala et al., 2006Motiwala AS, Li L, Kapur V et al. (2006) Current understanding of the genetic diversity of Mycobacterium avium subsp. paratuberculosis. Microbes Infect 8:1406–1418.; Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.). Moreover, it has the potential to be applied directly to clinical samples and does not require highly specialised personnel or sophisticated equipment. Consequently, this method is particularly desirable for MAP because of the very slow-growing nature of these organisms (Bull et al., 2003Bull TJ, Sidi-Boumedine K, McMinn EJ et al. (2003) Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol Cell Probes 17:157–164.) and could be easily applied in low-income countries. In this study, we aimed to study MAP strain diversity in Argentina by means of MIRU-VNTR.
Materials and Methods
MAP isolates and DNA extraction
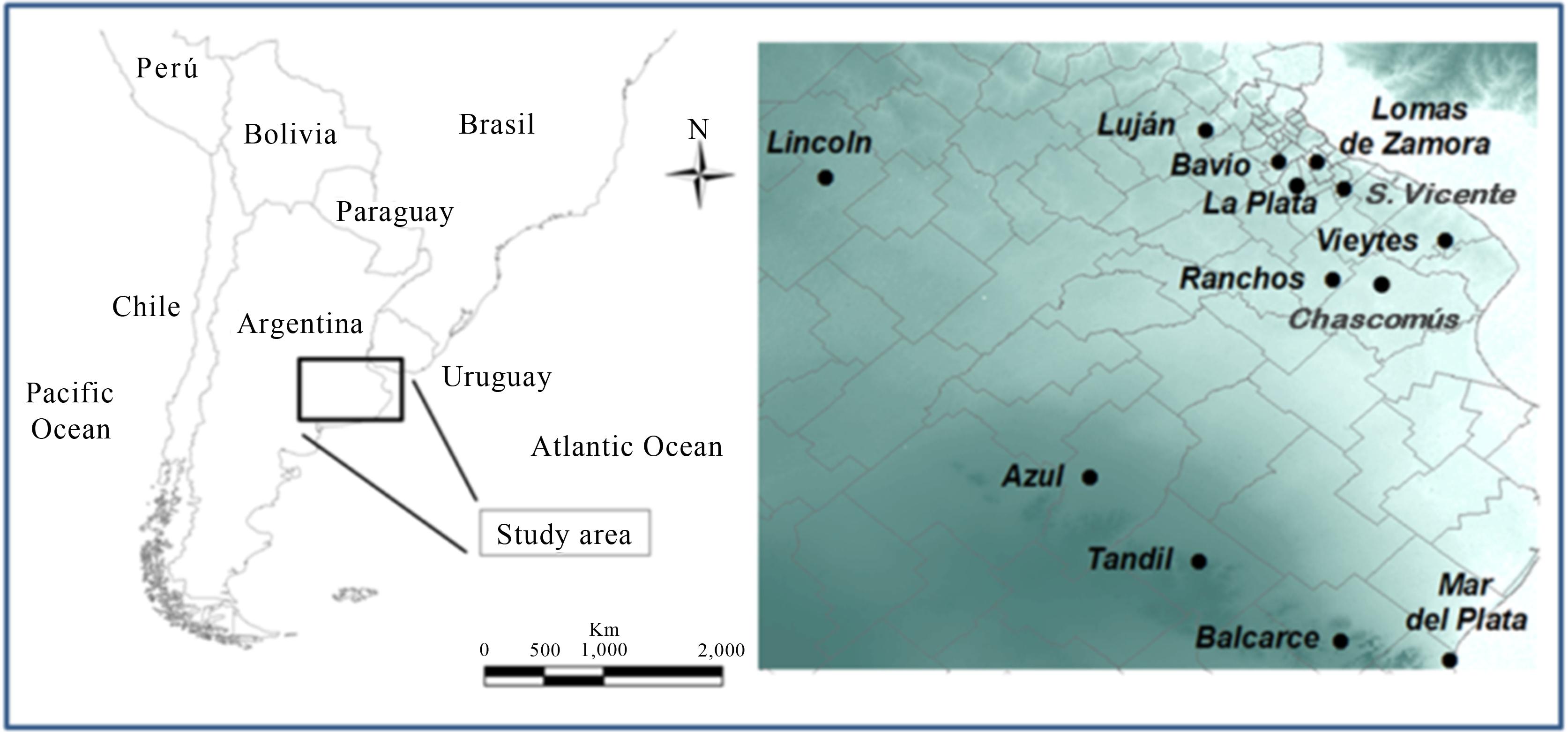
We tested ninety-seven isolates from a MAP collection, as confirmed by their mycobactin J dependence for growth, IS900-PCR (Englund et al., 1999Englund S, Ballagi-Pordany A, Bölske G et al. (1999) Single PCR and nested PCR with a mimic molecule for detection of Mycobacterium avium subsp. paratuberculosis. Diagn Micr Infec Dis 33:163–171.) and F57-PCR (Vansnick et al., 2004Vansnick E, De Rijk P, Vercammen F et al. (2004) Newly developed primers for the detection of Mycobacterium avium subspecies paratuberculosis. Vet Microbiol 100:197–204.). These isolates were classified by PCR and restriction endonuclease analysis (PCR-REA) as "cattle type" (C type) (Marsh et al., 1999Marsh I, Whittington R, Cousins D. (1999) PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311. Mol Cell Probes 13:115–126.). The tested clinical isolates corresponded to cattle, (n = 78) goats (n = 16) and sheep (n = 3) belonging to herds (n = 23) from thirteen geographic localities of the Buenos Aires province (Fig. 1). The isolates were obtained over the period from 2005 to 2010. A loopful of each isolate was suspended in sterile distilled water, and cell lysis was performed by serial freeze-boiling cycles. The samples were centrifuged (10,000 × g, 5 min), and 2 μL of the supernatant was used as the template in PCR reactions.
MIRU-VNTR typing: PCR conditions and number of tandem repeat determination
MIRU-VNTR typing was used for eight different MAP-specific markers, as described by Thibault et al., and designated VNTR292, MIRUX3 (alias MIRU2 and MIRU3, respectively, Bull et al., 2003Bull TJ, Sidi-Boumedine K, McMinn EJ et al. (2003) Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol Cell Probes 17:157–164.; Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.) VNTR25, VNTR47, VNTR3, VNTR7, VNTR10 and VNTR32 (Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.). The primers and PCR conditions were as suggested previously (Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.), with minor modifications. The amplification mixture consisted of 1X buffer (10 mM Tris-HCl pH 9; 50 mM KCl and 0.1% Triton X-100, 2.5 mM MgCl2) 0.2 mM each dNTP, 1 μM each primer and 1.25 U GoTaq polymerase (Promega). MgCl2 was supplemented for MIRUX3 (2 μL/reaction), and DMSO and betaine were supplemented when amplifying VNTR loci 47, 3, 7, 10 and 32, as suggested previously. The described previously annealing temperatures were employed, except for locus 47, for which the annealing temperature during the first 10 cycles was decreased 1 °C per cycle from 69 °C to 59 °C, according to a touch-down protocol, and then a final annealing temperature at 64 °C was selected for 35 cycles. The PCR products were resolved by 3.5% agarose gel electrophoresis, and the molecular weights were determined using 100 bp and 50 bp DNA markers (Promega Corp.) and BioNumerics software (Applied Maths, Belgium). The number of tandem repeats in each locus was assigned according to a previous report (Radomski et al., 2010Radomski N, Thibault VC, Karoui C et al. (2010) Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol 48:1026–1034.; see Supplementary tables). Numerical profiles were classified according to INMV (INRA, Nouzilly, MIRU-VNTR) combinations, as suggested previously (Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.; Radomski et al., 2010Radomski N, Thibault VC, Karoui C et al. (2010) Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol 48:1026–1034.). DNA from M. avium subsp. paratuberculosis K10 was included as a control (INMV2).
Index of discrimination
The allelic diversity for each locus and the discriminatory power of complete MIRU-VNTR scheme typing were determined using the Hunter and Gaston discriminatory index (Hunter, 1990Hunter P (1990) Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol 28:1903–1905.; Hunter and Gaston, 1988Hunter PR, Gaston MA (1988) Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466.) calculated using Simpson's index of diversity formula, as follows:
where D corresponds to the index of discriminatory power, N to the number of unrelated strains tested, S to the number of different types, and xj to the number of strains belonging to the jth type, assuming that strains will be classified into mutually exclusive categories.
This index was estimated considering epidemiologically unrelated strains and was calculated using the on-line software: http://insilico.ehu.es/mini_tools/discriminatory_power/, University of the Basque Country.
Epidemiological study
The eBURST algorithm described by Feil et al. (2004)Feil EJ, Li BC, Aanensen DM et al. (2004) eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530. was initially developed for MLST but is also suitable for MLVA studies. The goe-BURST algorithm (goeburst.phyloviz.net/) (Francisco et al., 2009Francisco AP, Bugalho M, Ramírez M et al. (2009) Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics doi:10.1186/1471-2105-10-152.
https://doi.org/10.1186/1471-2105-10-152...
), which uses the same clustering rules as eBURST (Feil et al., 2004Feil EJ, Li BC, Aanensen DM et al. (2004) eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530.) but provides a global optimal solution, was used to determine relationships between the obtained profiles. Clonal complexes were defined as MIRU-VNTR loci that are linked through single-locus variants (SLVs) and named on the basis of the predicted founder, which is the MIRU-VNTR pattern associated with most SLVs.
Results
MIRU-VNTR genotypes: Frequency, hosts and geographic distribution
The overall analysis yielded seven different INMV MIRU-VNTR patterns in the MAP isolates studied (n = 97). The INMV1 pattern grouped 65% of the isolates and was the most frequent. The rest of the genotypes were less represented, with INMV2 (11.3%), INMV16 (8.2%), and INMV33 (7.2%) being almost equally frequent. Although we were not able to amplify loci 292 and 7 in three isolates, the other loci indicated a pattern different from those mentioned above. This incomplete pattern represented 3.1% of the isolates. The most frequent patterns (INMV1 and INMV2) were described in at least two hosts (cattle, goat and sheep or cattle and goat, respectively), whereas the less frequent ones (INMV 11, INMV13, INMV16, INMV33, INMVx) were related to cattle (Table 1). The most common MIRU-VNTR patterns were widely distributed geographically, which limits their usefulness for tracing the geographic spread of infection. The MAP isolates from San Vicente showed the pattern INMV 13, and this pattern occurs exclusively in that location. The patterns INMV 11, INMV 16 and INMV 33 were exclusively from Tandil (Table 1 and Fig. 2).
GoeBURST clustering of the seven MIRU-VNTR profiles. Locations are represented in different colours. Differential loci between the patterns are shown in black numbers. The size of the pie is related to the number of samples.
Analysis of profiles by individual herds
Those herds with more than one isolate were considered for studying the coexistence of different genotypes. We found more than one strain in five out of nine herds. The herd with the highest number of isolates studied (Herd I, Tandil, n = 39) showed the highest diversity (five patterns) (Table 2). Moreover, two isolates were analysed from one animal of this herd, and two patterns were confirmed (not shown). The patterns found (INMV1 and INMV16) were the most frequent in this herd.
Allelic diversity and discriminatory power
Thirty-two non-epidemiologically related isolates were resampled to carry out both allelic diversity and discriminatory power analyses. According to our study, VNTR292 showed the maximum allelic diversity for Argentinean MAP strains (0.521). In contrast, VNTR loci 3, 10 and 32 were un-informative. The discriminatory power of the remaining MIRU-VNTR loci (X3, 25, 47 and 7) was weak, similar to most previous studies (Table 3). With the exception of VNTR32, which has eight tandem repeats (TRs), the rest of the loci showed a TR copy number lower than five. MIRU X3 was the only locus with one tandem repeat, and alleles without TRs were absent in the studied isolates. The complete MIRU-VNTR scheme discriminatory index D reached 0.6331. However, the discriminatory index was unchanged when the number of MIRU-VNTR loci was reduced to VNTR292-MIRUX3-VNTR25-VNTR47 and VNTR7, and the exclusion of VNTR47 or VNTR25 from the last combination did not reflect a significant difference in the discriminatory power reached (0.6086). Therefore, these findings indicate that a reduced MIRU-VNTR scheme (VNTR292-MIRUX3-VNTRs25/47 and VNTR7) yields discriminatory values comparable to the scheme of the complete 8 loci.
Frequency of tandem repeats in each MIRU-VNTR locus and comparative allelic diversity between different studies.
goeBURST analysis
To study the relationship among MIRU-VNTR patterns, we performed a cluster analysis using the goeBURST algorithm. The goeBURST clustering suggested pattern INMV2 as the primary founder. This pattern, together with INMV1, was the most geographically widespread, and only these two patterns were isolated from different hosts (Figure 2 and Table 1).
Discussion
The import of live animals from the United Kingdom is considered the most probable route of the introduction of M. bovis to Argentina, and this fact is supported by the genotyping of isolates from both regions (Cataldi et al., 2002Cataldi AA, Gioffré A, Santángelo MP et al. (2002) The genotype of the principal Mycobacterium bovis in Argentina is also that of the British isles: did bovine tuberculosis come from Great Britain? Rev Argent Microbiol 34:1–6.; Smith et al., 2011Smith NH, Berg S, Dale J et al. (2011) European 1: a globally important clonal complex of Mycobacterium bovis. Infect Genet Evol 11:1340–1351.). Considering the nature of MAP, we hypothesised that the genotypes/patterns of MAP from our country and Europe are also common. Our results expand an earlier study that examined the diversity of Argentinean MAP isolates using IS900-RFLP (Moreira et al., 1999Moreira AR, Paolicchi F, Morsella C et al. (1999) Distribution of IS900 restriction fragment length polymorphism types among animal Mycobacterium avium subsp. paratuberculosis isolates from Argentina and Europe. Vet Microbiol 70: 251–259.). MIRU-VNTR is faster and less labour-intensive than RFLP and has been internationally adopted (Stevenson et al., 2009Stevenson K, Alvarez J, Bakker D et al. (2009) Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol doi: 10.1186/1471-2180-9-212.
https://doi.org/10.1186/1471-2180-9-212...
, Inagaki et al., 2009Inagaki T, Nishimori K, Yagi T et al. (2009) Comparison of a variable-number tandem-repeat (VNTR) method for typing Mycobacterium avium with mycobacterial interspersed repetitive-unit-VNTR and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 47:2156–2164., Castellanos et al., 2010Castellanos E, Romero B, Rodríguez S et al. (2010) Molecular characterization of Mycobacterium avium subspecies paratuberculosis Type II and III isolates by a combination of MIRU-VNTR loci. Vet Microbiol 144:118–126.; Radomski et al., 2010Radomski N, Thibault VC, Karoui C et al. (2010) Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol 48:1026–1034.; Fernández-Silva et al., 2011Fernández-Silva JA, Abdulmawjood A, Bülte M (2011) Diagnosis and molecular characterization of Mycobacterium avium subsp. paratuberculosis from dairy cows in Colombia. Vet Med Int doi:10.4061/2011/352561.
https://doi.org/10.4061/2011/352561...
, Biet et al., 2012Biet F, Sevilla IA, Cochard T et al. (2012) Inter- and intra-subtype genotypic differences that differentiate Mycobacterium avium subspecies paratuberculosis strains. BMC Microbiol 12:264.). Hence, we selected a previously reported MIRU-VNTR scheme to study the diversity of circulating strains in Argentina in an effort to eventually perform a comparative analysis between different locations. We studied a larger sample size from three different hosts, covering a broad area of study compared to the aforementioned study. We demonstrate here that the most frequent MIRU-VNTR pattern is INMV1 (65%). This result is concordant with a previous report in which 51% of European isolates were typed as INMV1, which was also shown to be distributed in 6 out of the 7 countries assessed in that study (Stevenson et al., 2009Stevenson K, Alvarez J, Bakker D et al. (2009) Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol doi: 10.1186/1471-2180-9-212.
https://doi.org/10.1186/1471-2180-9-212...
). Additionally, patterns INMV1 and INMV2 were found to represent most of the isolates (36% and 34%, respectively) from 10 different countries and hosts (Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.). The goeBurst analysis indicated INMV2 as the founder genotype in our study. A similar result was obtained by the minimum-spanning-tree approach performed in a strain panel containing mainly European isolates (Radomski et al., 2010Radomski N, Thibault VC, Karoui C et al. (2010) Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol 48:1026–1034.).
To date, the patterns INMV1, INMV2, INMV11, INMV13, INMV16, INMV33 (this work), and INMV8 (Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.) have been described in Argentinean isolates. The currently available information for pattern distribution in the United Kingdom shows the presence of INMV1, INMV2, INMV19, INMV21, INMV25 (Stevenson et al., 2009Stevenson K, Alvarez J, Bakker D et al. (2009) Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol doi: 10.1186/1471-2180-9-212.
https://doi.org/10.1186/1471-2180-9-212...
) and INMV14 and INMV33 (Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.) in different hosts. Considering these data in combination with the fact that INMV1 and INMV2 are distributed worldwide, we cannot assume or discard the epidemiological link between the UK and Argentina using this VNTR-MIRU typing approach, and further evidence is required to confirm this hypothesis. Nonetheless, almost all described patterns in our study have been previously reported in European isolates (Thibault et al., 2007Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.). The combination of other typing technique(s) with MIRU-VNTR analysis has enabled an improvement of the discriminatory power (Thibault et al., 2008Thibault VC, Grayon M, Boschiroli ML et al. (2008) Combined multilocus short-sequence-repeat and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing of Mycobacterium avium subsp. paratuberculosis isolates. J Clin Microbiol 46:4091–4094.; Stevenson et al., 2009Stevenson K, Alvarez J, Bakker D et al. (2009) Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol doi: 10.1186/1471-2180-9-212.
https://doi.org/10.1186/1471-2180-9-212...
). More recently, Fernández Silva et al. were able to improve the discrimination between six INMV1 Argentinian strains, which were resolved into three genotypes using multilocus short sequence repeats (Fernández Silva et al., 2012Fernández-Silva JA, Abdulmawjood A, Akineden Ö et al. (2012). Genotypes of Mycobacterium avium subsp. paratuberculosis from South American countries determined by two methods based on genomic repetitive sequences. Trop Anim Health Prod 44:1123–1126.). Hence, the use of more than one typing method to further discriminate between isolates and the inclusion of a larger number of strains by random sampling could be more informative about the real epidemiological scenario in Argentina.
We found that the most important limitation of MIRU-VNTR typing was the lack of amplification for two loci. We herein report INMVx as an incomplete pattern because two MIRU-VNTR loci (292 and 7) remained refractory to amplification in three samples. This could be due to target sequence polymorphisms or to the absence of these loci in these strains.
The discriminatory index we obtained reached 0.63, in accordance with previous studies by Thibault et al. (2007)Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410. and Stevenson et al. (2009)Stevenson K, Alvarez J, Bakker D et al. (2009) Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol doi: 10.1186/1471-2180-9-212.
https://doi.org/10.1186/1471-2180-9-212...
, 0.75 and 0.64, respectively. Based on information derived from these previous studies and the present work, a reduced scheme can be considered for typing MAP isolates without a significant loss of discriminative power. In this regard, MIRU-VNTR loci 3, 10 and 32 can be excluded in future studies involving local strains, and moreover novel markers could be considered for improving the discriminatory power.
The coexistence of different genotypes within cattle herds has been previously reported (van Hulzen et al., 2011van Hulzen KJE, Heuven HCM, Nielen M et al. (2011) Different Mycobacterium avium subsp. paratuberculosis MIRU-VNTR patterns coexist within cattle herds. Vet Microbiol 148:419–424.; Fernández-Silva et al., 2011Fernández-Silva JA, Abdulmawjood A, Bülte M (2011) Diagnosis and molecular characterization of Mycobacterium avium subsp. paratuberculosis from dairy cows in Colombia. Vet Med Int doi:10.4061/2011/352561.
https://doi.org/10.4061/2011/352561...
). Within this context, cattle herd I (Tandil) with at least five different patterns constitutes a good example. The observed genotype co-existence within herds strongly suggests that a lack of animal monitoring prior to their introduction into a herd may represent a factor that is key to the observed diversity. Accordingly, we cannot rule out the presence of more than one genotype in all the tested herds, and a wider sampling should be performed to examine this. For most of the herds assessed in this study, animals are currently introduced, or were introduced in the past, without prior testing, justifying this practice based on the apparent healthy condition of the animals. One case was the introduction of a bull for breeding purposes in an infected herd (herd II, San Vicente). The isolate from this animal showed a pattern different from those described for other isolates from the same herd. Considering that animals usually become infected early in their life, one can assume that this bull was already infected. However, we also studied a MAP-infected flock (herd VI, Mar del Plata) in which the PTB seroprevalence was 48% (Fiorentino et al., 2012Fiorentino MA, Gioffré A, Cirone K et al. (2012) First isolation of Mycobacterium avium subsp. paratuberculosis in a dairy goat in Argentina: Pathology and molecular characterization. Small Ruminant Res http://dx.doi.org/10.1016/j.smallrumres.2012.06.010.
http://dx.doi.org/10.1016/j.smallrumres....
). In this case, the introduction of foreign animals was not a common practice, which clearly correlates with the presence of isolates sharing the same pattern. It is important to mention that, unfortunately, this MAP-infected herd is a frequent supplier of animals to neighbouring herds.
Since the early epidemiological studies on PTB, molecular typing techniques have been described and applied. The MIRU-VNTR approach has been used to type M. avium complex strains from different countries, such as France, Germany, Czech Republic, Finland, Greece, The Netherlands, Norway, Scotland, Spain, Japan and more recently Colombia and Argentina (Möbius et al., 2008Möbius P, Luyven G, Hotzel H et al. (2008) High genetic diversity among Mycobacterium avium subsp. paratuberculosis strains from German cattle herds shown by combination of IS900 restriction fragment length polymorphism analysis and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing. J Clin Microbiol 46:972–981.; Stevenson et al., 2009Stevenson K, Alvarez J, Bakker D et al. (2009) Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol doi: 10.1186/1471-2180-9-212.
https://doi.org/10.1186/1471-2180-9-212...
; Inagaki et al., 2009Inagaki T, Nishimori K, Yagi T et al. (2009) Comparison of a variable-number tandem-repeat (VNTR) method for typing Mycobacterium avium with mycobacterial interspersed repetitive-unit-VNTR and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 47:2156–2164.; Castellanos et al., 2010Castellanos E, Romero B, Rodríguez S et al. (2010) Molecular characterization of Mycobacterium avium subspecies paratuberculosis Type II and III isolates by a combination of MIRU-VNTR loci. Vet Microbiol 144:118–126.; Radomski et al., 2010Radomski N, Thibault VC, Karoui C et al. (2010) Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol 48:1026–1034.; Fernández-Silva et al., 2011Fernández-Silva JA, Abdulmawjood A, Bülte M (2011) Diagnosis and molecular characterization of Mycobacterium avium subsp. paratuberculosis from dairy cows in Colombia. Vet Med Int doi:10.4061/2011/352561.
https://doi.org/10.4061/2011/352561...
, this work). Despite the high acceptance of this technique, the results from different studies are often difficult to compare due to the lack of standardisation. At present, a consensus on which is the best technique or combination of multiple genotyping techniques is urgently needed to definitively understand the global epidemiology of PTB, and this can be achieved through collaborative work. This report constitutes the first MIRU-VNTR study of a large panel of strains from one of the most productive regions of Argentina.
Acknowledgments
This work was partially supported by grants from INTA (AESA202831) and BiotecSur. The authors acknowledge V. Rocha and V. Montenegro for technical assistance, Dr. A. Bernardelli and L. Fernández for bibliographic support and Dr. J. Sabio-García for critical reading of the manuscript. AG, PR and MIR are members of the Research Career of the CONICET, Buenos Aires, Argentina.
References
- Attili AR, NguNgwa V, Preziuso S et al. (2011) Ovine paratuberculosis: A seroprevalence study in dairy flocks reared in the Marche region, Italy. Vet Med Int doi:10.4061/2011/782875.
» https://doi.org/10.4061/2011/782875 - Beard PM, Daniels MJ, Henderson D et al. (2001) Paratuberculosis infection of nonruminant wildlife in Scotland. J Clin Microbiol 39:1517–1521.
- Bernardelli A, Cicuta NE, Nicola A et al. (2002) Ovine paratuberculosis: field diagnosis. 7th International Colloquium on Paratuberculosis Bilbao, Spain.
- Bernardelli A, Zumárraga M, Alonso B et al. (2005) Paratuberculosis in Mouflon (Ovis musimon). 8th International Colloquium on Paratuberculosis, Copenhagen, Denmark.
- Biet F, Sevilla IA, Cochard T et al. (2012) Inter- and intra-subtype genotypic differences that differentiate Mycobacterium avium subspecies paratuberculosis strains. BMC Microbiol 12:264.
- Buergelt CD, Layton AW, Ginn PE et al. (2000) The pathology of spontaneous paratuberculosis in the North American bison (Bison bison). Vet Pathol 37:428–438.
- Bull TJ, Sidi-Boumedine K, McMinn EJ et al. (2003) Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol Cell Probes 17:157–164.
- Castellanos E, Romero B, Rodríguez S et al. (2010) Molecular characterization of Mycobacterium avium subspecies paratuberculosis Type II and III isolates by a combination of MIRU-VNTR loci. Vet Microbiol 144:118–126.
- Cataldi AA, Gioffré A, Santángelo MP et al. (2002) The genotype of the principal Mycobacterium bovis in Argentina is also that of the British isles: did bovine tuberculosis come from Great Britain? Rev Argent Microbiol 34:1–6.
- Cook KL, Britt JS, Bolster CH (2010) Survival of Mycobacterium avium subsp. paratuberculosis in biofilms on livestock watering trough materials. Vet Microbiol 141:103–109.
- Djønne B, Pavlik I, Svastova P et al. (2005) IS900 Restriction Fragment Length Polymorphism (RFLP) Analysis of Mycobacterium avium subsp. paratuberculosis isolates from goats and cattle in Norway. Acta Vet Scand 46:13–18.
- Englund S, Ballagi-Pordany A, Bölske G et al. (1999) Single PCR and nested PCR with a mimic molecule for detection of Mycobacterium avium subsp. paratuberculosis Diagn Micr Infec Dis 33:163–171.
- Feil EJ, Li BC, Aanensen DM et al. (2004) eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530.
- Fernández-Silva JA, Abdulmawjood A, Bülte M (2011) Diagnosis and molecular characterization of Mycobacterium avium subsp. paratuberculosis from dairy cows in Colombia. Vet Med Int doi:10.4061/2011/352561.
» https://doi.org/10.4061/2011/352561 - Fernández-Silva JA, Abdulmawjood A, Akineden Ö et al. (2012). Genotypes of Mycobacterium avium subsp. paratuberculosis from South American countries determined by two methods based on genomic repetitive sequences. Trop Anim Health Prod 44:1123–1126.
- Fiorentino MA, Gioffré A, Cirone K et al. (2012) First isolation of Mycobacterium avium subsp. paratuberculosis in a dairy goat in Argentina: Pathology and molecular characterization. Small Ruminant Res http://dx.doi.org/10.1016/j.smallrumres.2012.06.010
» http://dx.doi.org/10.1016/j.smallrumres.2012.06.010 - Francisco AP, Bugalho M, Ramírez M et al. (2009) Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics doi:10.1186/1471-2105-10-152.
» https://doi.org/10.1186/1471-2105-10-152 - Ghosh P, Hsu C, Alyamani EJ et al. (2012) Genome-wide analysis of the emerging infection with Mycobacterium avium subspecies paratuberculosis in the Arabian camels (Camelus dromedarius). PLoS One doi: 10.1371/journal.pone.0031947.
» https://doi.org/10.1371/journal.pone.0031947 - Hunter PR, Gaston MA (1988) Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466.
- Hunter P (1990) Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol 28:1903–1905.
- Inagaki T, Nishimori K, Yagi T et al. (2009) Comparison of a variable-number tandem-repeat (VNTR) method for typing Mycobacterium avium with mycobacterial interspersed repetitive-unit-VNTR and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 47:2156–2164.
- Jorge MC, Schettino DM, Torres P et al. (2000) Primera descripción de infección concomitante de tuberculosis y paratuberculosis en ovinos lecheros en Argentina. Rev Sci Tech Off Int Epizoot 19:800–809.
- Marsh I, Whittington R, Cousins D. (1999) PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311 Mol Cell Probes 13:115–126.
- Möbius P, Luyven G, Hotzel H et al. (2008) High genetic diversity among Mycobacterium avium subsp. paratuberculosis strains from German cattle herds shown by combination of IS900 restriction fragment length polymorphism analysis and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing. J Clin Microbiol 46:972–981.
- Moreira AR, Paolicchi F, Morsella C et al. (1999) Distribution of IS900 restriction fragment length polymorphism types among animal Mycobacterium avium subsp. paratuberculosis isolates from Argentina and Europe. Vet Microbiol 70: 251–259.
- Motiwala AS, Li L, Kapur V et al. (2006) Current understanding of the genetic diversity of Mycobacterium avium subsp. paratuberculosis Microbes Infect 8:1406–1418.
- Mura M, Bull TJ, Evans H et al. (2006) Replication and long-term persistence of bovine and human strains of Mycobacterium avium subsp. paratuberculosis within Acanthamoeba polyphaga Appl Environ Microbiol 72:854–859.
- Nader A, Bernardelli A (1986) Utilización del test tuberculínico intradérmico comparativo en ovinos como prueba de screening en paratuberculosis. Revista de Medicina Veterinaria 67:335–338.
- OIE. 2009. World Animal Health Information Database (WAHID) - Version: 1.4 Available at http://www.oie.int
» http://www.oie.int - Paolicchi FA, Vagnozzi A, Morsella CG et al. (2001) Paratuberculosis in red deer (Cervus elaphus): an immunohistochemical study. J Vet Med, Series B 48:313–320.
- Paolicchi F, Zumárraga J, Gioffré A et al. (2003) Application of different methods for the diagnosis of paratuberculosis in a dairy cattle herd in Argentina. J Vet Med 50:20–26.
- Pickup RW, Rhodes G, Arnott S et al. (2005) Mycobacterium avium subsp. paratuberculosis in the catchment area and water of the river Taff in South Wales, United Kingdom, and its potential relationship to clustering of Crohn's disease cases in the city of Cardiff. Appl Environ Microbiol 71:2130–2139.
- Radomski N, Thibault VC, Karoui C et al. (2010) Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J Clin Microbiol 48:1026–1034.
- Salgado M, Herthnek D, Bölske G et al. (2009) First isolation of Mycobacterium avium subsp. paratuberculosis from wild guanacos (Lama guanicoe) on Tierra del Fuego Island. J Wildl Dis 45:295–301.
- Smith NH, Berg S, Dale J et al. (2011) European 1: a globally important clonal complex of Mycobacterium bovis Infect Genet Evol 11:1340–1351.
- Stevenson K, Alvarez J, Bakker D et al. (2009) Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol doi: 10.1186/1471-2180-9-212.
» https://doi.org/10.1186/1471-2180-9-212 - Thibault VC, Grayon M, Boschiroli ML et al. (2007) New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410.
- Thibault VC, Grayon M, Boschiroli ML et al. (2008) Combined multilocus short-sequence-repeat and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing of Mycobacterium avium subsp. paratuberculosis isolates. J Clin Microbiol 46:4091–4094.
- van Hulzen KJE, Heuven HCM, Nielen M et al. (2011) Different Mycobacterium avium subsp. paratuberculosis MIRU-VNTR patterns coexist within cattle herds. Vet Microbiol 148:419–424.
- Vansnick E, De Rijk P, Vercammen F et al. (2004) Newly developed primers for the detection of Mycobacterium avium subspecies paratuberculosis Vet Microbiol 100:197–204.
- Verna A, Morsella C, Zumárraga M et al. (2002) Strains characterization by PCR and RFLP from Mycobacterium avium subsp. paratuberculosis isolates of red deer with paratuberculosis. 7th International Colloquium on Paratuberculosis, Bilbao, Spain.
- Whittington RJ, Marshall DJ, Nicholls PJ et al. (2004) Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl Environ Microbiol 70:2989–3004.
Publication Dates
-
Publication in this collection
Apr-Jun 2015
History
-
Received
03 Apr 2014 -
Accepted
16 Nov 2014