Abstracts
Euglossine bees interact with more than 60 plant families of the Neotropical region. The richness and abundance of these bees have been intensively studied in different ecosystems using the methodology of capturing males with chemical baits. Females are poorly known for most of the species and morphological characters for their taxonomic classification have not yet been described. The purpose of this study was to use allozymes and restriction patterns of the mitochondrial regions 16S and Cyt b to identify species of Euglossa Latreille. Bees were collected while visiting Thevetia peruviana (Apocynaceae) flowers in five cities of the state of São Paulo, Brazil. Three Euglossa species were identified among the 305 individuals collected. Euglossa cordata (L.) was the only species found in all cities, while E. securigera Dressler and E. townsendi Cockerell were restricted to two and one cities respectively. EST-3 was a diagnostic marker, whereas ICD, MDH, ME and PGM were informative for species identification when used in combination. Restriction by VspI of the amplified 16S fragment differentiated the three species and showed intraspecific polymorphism for E. cordata and E. securigera. The Cyt b region showed distinctive patterns for E. townsendi but it was not possible to differentiate the other two species. Our results describe potentially useful genetic markers for the identification of Euglossa spp. at the species and group level.
Orchid bee; urban area; Thevetia peruviana; allozyme; PCR-RFLP
As abelhas euglossíneas interagem com mais de 60 famílias de plantas da Região Neotropical. Sua riqueza e abundância têm sido intensamente estudadas em diferentes ecossistemas utilizando-se a metodologia de captura de machos em iscas-armadilhas. As fêmeas, entretanto, são pouco conhecidas para a maioria das espécies, e caracteres morfológicos que permitam sua identificação taxonômica não têm sido descritos. O propósito deste trabalho foi utilizar alozimas e padrões de restrição das regiões mitocondriais 16S e Cit b para identificar espécies do gênero Euglossa Latreille. As abelhas foram coletadas enquanto visitavam flores de Thevetia peruviana (Apocynaceae) em cinco cidades do estado de São Paulo. Foram identificadas três espécies de Euglossa entre os 305 indivíduos coletados. Euglossa cordata (L.) foi a única espécie presente em todas as cidades, enquanto E. securigera Dressler e E. townsendi Cockerell foram encontradas em duas e uma cidade, respectivamente. EST-3 mostrou-se ser marcador diagnóstico, enquanto ICD, MDH, ME e PGM foram locos informativos para a identificação de espécies quando considerados conjuntamente. A restrição com VspI da região 16S amplificada, além de diferenciar as três espécies, apresentou polimorfismo intraespecífico para E. cordata e E. securigera. A região Cit b apresentou padrões característicos para E. townsendi, mas não permitiu diferenciar as outras duas espécies. Os resultados descrevem marcadores genéticos potencialmente úteis para a identificação de Euglossa spp. ao nível de espécie e grupo de espécies.
Abelha das orquídeas; área urbana; Thevetia peruviana; alozima; PCR-RFLP
SYSTEMATICS, MORPHOLOGY AND PHYSIOLOGY
Molecular identification of species of the Genus Euglossa Latreille (Hymenoptera: Apidae, Euglossini)
Identificação molecular de espécies do gênero Euglossa Latreille (Hymenoptera: Apidae, Euglossini)
Margarita M. López-UribeI, II; Marco A. Del LamaI, III
ILab. Genética Evolutiva de Himenópteros, Depto. Genética e Evolução, Univ. Federal de São Carlos, Rodovia Washington Luis, km 235, 13565.905, São Carlos, SP, Brazil
IICurrent address: Dept. Entomology, Comstock Hall 3126 Cornell University, Ithaca, NY 14853-0901, USA, mml82@cornell.edu
ABSTRACT
Euglossine bees interact with more than 60 plant families of the Neotropical region. The richness and abundance of these bees have been intensively studied in different ecosystems using the methodology of capturing males with chemical baits. Females are poorly known for most of the species and morphological characters for their taxonomic classification have not yet been described. The purpose of this study was to use allozymes and restriction patterns of the mitochondrial regions 16S and Cyt b to identify species of Euglossa Latreille. Bees were collected while visiting Thevetia peruviana (Apocynaceae) flowers in five cities of the state of São Paulo, Brazil. Three Euglossa species were identified among the 305 individuals collected. Euglossa cordata (L.) was the only species found in all cities, while E. securigera Dressler and E. townsendi Cockerell were restricted to two and one cities respectively. EST-3 was a diagnostic marker, whereas ICD, MDH, ME and PGM were informative for species identification when used in combination. Restriction by VspI of the amplified 16S fragment differentiated the three species and showed intraspecific polymorphism for E. cordata and E. securigera. The Cyt b region showed distinctive patterns for E. townsendi but it was not possible to differentiate the other two species. Our results describe potentially useful genetic markers for the identification of Euglossa spp. at the species and group level.
Keywords: Orchid bee, urban area, Thevetia peruviana, allozyme, PCR-RFLP
RESUMO
As abelhas euglossíneas interagem com mais de 60 famílias de plantas da Região Neotropical. Sua riqueza e abundância têm sido intensamente estudadas em diferentes ecossistemas utilizando-se a metodologia de captura de machos em iscas-armadilhas. As fêmeas, entretanto, são pouco conhecidas para a maioria das espécies, e caracteres morfológicos que permitam sua identificação taxonômica não têm sido descritos. O propósito deste trabalho foi utilizar alozimas e padrões de restrição das regiões mitocondriais 16S e Cit b para identificar espécies do gênero Euglossa Latreille. As abelhas foram coletadas enquanto visitavam flores de Thevetia peruviana (Apocynaceae) em cinco cidades do estado de São Paulo. Foram identificadas três espécies de Euglossa entre os 305 indivíduos coletados. Euglossa cordata (L.) foi a única espécie presente em todas as cidades, enquanto E. securigera Dressler e E. townsendi Cockerell foram encontradas em duas e uma cidade, respectivamente. EST-3 mostrou-se ser marcador diagnóstico, enquanto ICD, MDH, ME e PGM foram locos informativos para a identificação de espécies quando considerados conjuntamente. A restrição com VspI da região 16S amplificada, além de diferenciar as três espécies, apresentou polimorfismo intraespecífico para E. cordata e E. securigera. A região Cit b apresentou padrões característicos para E. townsendi, mas não permitiu diferenciar as outras duas espécies. Os resultados descrevem marcadores genéticos potencialmente úteis para a identificação de Euglossa spp. ao nível de espécie e grupo de espécies.
Palabras-chave: Abelha das orquídeas, área urbana, Thevetia peruviana, alozima, PCR-RFLP
Euglossine bees interact with species from more than 60 plant families of the Neotropical forests and comprise almost 25% of the bee community in some ecosystems (Roubik & Hanson 2004). These bees, also known as orchid bees, are hard to detect in nature because of their fast flying, solitary habits and the difficulty of locating their nests (Cameron 2004). Knowledge on the biology of euglossine bees increased only after the isolation of compounds that mimic the fragrances males look for in flowers. As females are not attracted to these chemical baits, literature on systematics, ecology and genetics of orchid bees is mostly based on male data (Roubik & Ackerman 1987, Ramírez et al. 2002, Zayed et al. 2004).
Sampling restricted to chemical baits can underestimate abundance and richness of euglossine bees since some species are not attracted to the aromatic compounds frequently used (Ackerman 1983, Roubik & Ackerman 1987, Rebêlo & Garófalo 1997). Therefore, alternative sampling from flowers, nests and other chemical sources is necessary to obtain an accurate estimate of the number of species present in different ecosystems. Besides, the use of other sampling techniques would also provide knowledge about the behavior and ecological preferences of females of the Euglossini tribe.
Taxonomic identification of euglossine females may be problematic for some species, mainly for the most specious genera Euglossa Latreille and Eufriesea Cockerell (Ramírez et al. 2002). This difficulty generally arises because (1) descriptions of species have usually been based on male holotypes and (2) female individuals are rare in entomological collections (Bonilla-Gómez & Nates-Parra 1992). In particular, females of the genus Euglossa Latreille are unknown for over 40% of the species and morphological characters for their taxonomic identification have not yet been described.
Genetic markers have been intensively used as a tool for species identification when morphological characters are unknown or of difficult interpretation (Brunner et al. 2002, Naeole & Haymer 2003, Schama et al. 2005). Among the molecular markers, allozymes may be used as diagnostic markers for identification of cryptic species, as they have low intraspecific polymorphism levels and usually show fixed alleles within species (Schama et al. 2005). However, the need of fresh material for allozyme analysis restrains its large-scale use for systematic purposes. Since the development of the PCR (Polymerase Chain Reaction), DNA markers such as RAPD, AFLP, PCR-RFLP and DNA sequences have been commonly used for species identification since a small amount of dried material is sufficient for the analysis. Therefore, molecular markers are suitable alternative tools for the taxonomic identification of groups in challenging cases.
As female identification is a problematic issue for the genus Euglossa, the aim of this study was to use allozymes and restriction patterns of the mitochondrial regions 16S and Cyt b for the identification of Euglossa spp. Latreille. Specimens were collected while visiting Thevetia peruviana (Apocynaceae) flowers in five urban areas of the state of São Paulo, Brazil. These mitochondrial loci were tested because they were previously used for species identification in other groups (Gajardo et al. 2004, Ellis et al. 2006). We estimated (1) interspecific variation to identify diagnostic markers and (2) intraspecific variation to evaluate the accuracy of the markers proposed. The molecular methodology here described provides a reliable and inexpensive tool for identification of euglossine specimens that visit T. peruviana flowers in the urban areas sampled.
Material and Methods
Sample collection. Euglossine bees were collected in the urban areas of Rifaina (20º0450"S 47º2517"W), São Carlos (22º0016"S 47º5318"W), Pedregulho (20º1525"S 47º2836"W), Jaboticabal (21º1517"S 48º1920"W) and Araras (22º2125"S 47º2303"W), SP (Fig. 1), during the hot rainy season of 2003 to 2005. Specimens were collected in plastic bags when they were exiting T. peruviana flowers, placed in plastic vials and stored at -20ºC until analysis. As T. peruviana is an ornamental exotic plant, the number of sites sampled in each city varied among localities according to the abundance of plants in each area (Fig. 1). A total of 305 individuals of Euglossa, 277 females and 28 males, were captured (Table 1). Males were taxonomically identified, following the key of Rebêlo and Moure (1995), and later used as controls of the allozyme and mitochondrial restriction patterns for the identification of females.
Allozyme analysis. Allozymes were used to identify the number of species present in our samples. The "field for recombination" criterion was employed, identifying species based on the non-overlapping sets of heterozygous individuals (Sites & Marshall 2003). Proteins were extracted from the head in a 0.2% 2-mercaptoethanol solution and later analyzed through horizontal electrophoresis in 14% starch gels using (1) Tris Citric Acid pH 7.5, (2) Tris Citric Acid pH 8.0 and (3) Tris Citric Acid pH 8.0 Boric Acid pH 8.3 as buffers. Twenty-two enzymes corresponding to 24 loci were tested: acid phosphatase (ACP, TC 7.5); aconitase (ACO, TC 7.5); adenylate kinase (AK, TC 8.0); aldolase (ALD, TC 7.5); arginine kinase (ARGK, TC 8.0 and TCB 8.0-8.3); creatine kinase (CK, TC 8.0); esterase (EST TC 7.5 and TCB 8.0-8.3); fumarase (FUM, TC 7.5); glucose-6-phosphate dehydrogenase (G6PDH, TC 7.5); a-glycerophosphate dehydrogenase (GPDH, TC 8.0); glucosephosphate isomerase (GPI, TC 8.0); b-hidroxybutirate dehydrogenase (HBDH, TC 7.5); hexokinase (HK, TC 8.0); isocitric dehydrogenase (ICD, TC 7.5); leucine aminopeptidase (LAP, TC 8.0 and TCB 8.0-8.3); malate dehydrogenase (MDH, TC 8.0); malic enzyme (ME, TC 8.0); mannose-6-phosphate isomerase (MPI, TC 7.5); peptidase A (leu-ala) (PEP-A, TCB 8.0-8.3); phosphoglucomutase (PGM, TC 8.0); 6-phosphogluconate dehydrogenase (6-PGD, TC 7.5) and superoxide dismutase (SOD, TCB 8.0-8.3). All enzymatic reactions were prepared according to Harris and Hopkinson (1976).
PCR-RFLP analysis. Total genomic DNA was extracted from thoracic tissues grounded in a 1.5 ml microtube with extraction buffer and later incubated with proteinase K at 60ºC for 2h. Incubation was followed by a standard phenol-chloroform and ethanol precipitation protocol (Sheppard & McPheron 1991). The resultant DNA pellet was resuspended in 50 µl of TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA) and stored at -20ºC.
PCR products were amplified in a Bio-Rad Gene thermocycler. For amplification of the Cyt b fragment, we used the primers described by Crozier et al. (1991) with 30 amplification cycles (94ºC for 30 s; 56ºC for 15 s; 72ºC for 1 min). The 16S fragment was amplified using the primers 16SWb (Dowton & Austin 1994) and 874-16SlR (Cameron et al. 1992). PCR conditions consisted of 35 amplification cycles (94ºC for 45 s; 5 cycles at 46ºC, 5 cycles at 48ºC and 25 cycles at 50ºC for 45 s; 72ºC for 45 s) followed by a final elongation step for 5 min at 72ºC. Each reaction was performed in a 25 µl final volume containing 12.5 µM of each dNTP, 2.5 (Cyt b) or 5.0 (16S) mM of MgCl2, 2.5 µl Biotools Buffer 10x, 0.5 µM of each Primer, 0.8U of Taq Polymerase Biotools and 1 µl of DNA solution.
Digestion of the amplified DNA products was performed in a 10 µl final volume solution containing 1 µl of the PCR product, 1 µl of One-Phor-All (Amershan) buffer and 1 U of the restriction enzyme. Restriction patterns were visualized in 12% silver stained polyacrylamide gels. The endonucleases BglII, DraI, EcoRI, MboI, TaqI and VspI were initially tested for the Cyt b and 16S fragments because they show polymorphism when used in Apis mellifera L. for these regions.
After characterizing different restriction mitotypes with these endonucleases, fragments of each prior identified species were sequenced to check the restriction patterns visualized on the polyacrylamide gels and to detect possible new restriction polymorphisms. The amplified products of both regions were purified using 1 U of SAP (Shrimp Alkaline Phosphatase, Amersham Pharmacia Biotech) and 10 U of ExoI (Exonuclease I, Amersham Pharmacia Biotech) for 8 µl of DNA and later incubated at 37ºC for 1h and at 80ºC for 15 min. The sequencing reaction was performed in a 10 µl final volume reaction containing 3.5 µl of Save Money Buffer 2.5x, 0.5 µl of Big Dye (Applied Biosystems), 1 µM of Primer and 1 µl of DNA. Direct sequencing of forward and reverse strands was obtained in an ABI3700 automatic sequencer.
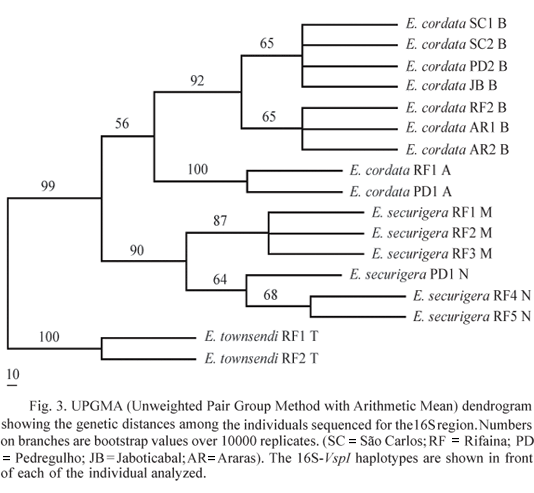
Data analysis. Sequences of the 16S and Cyt b regions were edited using CodonCode Aligner v 1.5.2 software (CodonCode, Dedham, Massachusetts, United States) and later on aligned using the CLUSTAL X multiple sequences editor (Thomson et al. 1997). Intraspecific polymorphism was estimated as the average number of pairwise differences within species (dW) and as nucleotide diversity (p, Nei 1987) using the ARLEQUIN 2.0 software (Schneider et al. 2000). Interspecific variation was estimated as the average number of pairwise differences between populations (dB) and as FST statistics (Weir & Cockerham 1984). To estimate the genetic similarity among sequences, a UPGMA (Unweighted Pair Group Method with Arithmetic Mean) genetic distance tree was constructed by the PAUP* program (Swofford 1999), resampling 10000 times to obtain bootstrap support values for each cluster. This method of tree construction was chosen because it is based on the overall similarity among sequences without assuming an evolutionary model. Sequences were analyzed with the Sequence Analysis 1.6.0 software (http://informagen.com/SA/) to assess restriction patterns with restriction enzymes.
Results
Species identification. The males captured were taxonomically identified as E. cordata (L.), E. securigera Dressler and E. townsendi Cockerell. Even though males were intended to be collected at all cities, 90% of the flower visitors were females, and consequently males were not sampled at all localities (Table 1). This preliminary classification could have underestimated the number of species because males were underrepresented in our samples; however, molecular analysis confirmed the presence of just three Euglossa species. E. cordata was the only species found at all localities and was also the most abundant. Samples from São Carlos, Jaboticabal and Araras were exclusively composed of E. cordata. E. securigera occurred in Rifaina and Pedregulho, while E. townsendi was found exclusively in Rifaina.
Allozyme analysis. Five of the 24 allozyme loci analyzed were informative for species identification. EST-3 was a diagnostic locus since it showed exclusive fixed alleles for each of the three species (Table 2). MDH, ME, ICD and PGM were also informative for species identification because they presented fixed alleles that, in association, were also diagnostic. Loci ACP, EST-1, HK-2, G6PDH and FUM showed intraspecific variation uninformative for species identification but useful for population genetic studies.
PCR-RFLP analysis. Three of the six endonucleases tested revealed variation for both mitochondrial regions: DraI, MboI and VspI. The restriction analysis of the 16S fragment with VspI showed diagnostic restriction patterns, allowing the identification of each of the three species. These 16S-VspI patterns varied in the number of restriction sites from seven to ten. However, some fragments were not visualized in the polyacrylamide gel because of their small size (Fig. 2). The largest fragment was characteristic of E. securigera (174) while the smallest fragments differentiated E. townsendi (34) from E. cordata (30). Besides being diagnostic for the three species, VspI digestion of the 16S fragment revealed two different mitotypes for E. cordata (CA; CB) and E. securigera (SM; SN) (Fig. 2). DraI also revealed intraspecific polymorphism in the 16S fragment, but was not useful for species identification, as E. cordata and E. securigera shared one pattern.
Restriction analyses of the Cyt b fragment with MboI and VspI revealed common patterns for E. cordata and E. securigera. However, E. townsendi was easily identified when the amplified fragment was digested with these restriction enzymes. The Cyt b fragment showed the occurrence of three MboI restriction sites for E. townsendi, whereas E. cordata and E. securigera presented four. Using VspI, one restriction site was detected for E. townsendi, whereas the other two species had at least two. VspI also revealed intraspecific polymorphism for E. cordata (CE; CF) but the CF pattern was shared by E. securigera.
Nucleotide sequences. All restriction patterns were confirmed by sequencing fragments of the 16S and Cyt b regions from at least five individuals of the three previously detected species. Sequences of the 16S region revealed a high number of polymorphic sites as well as fragments that were variable in size, among and within species, ranging from 579 bp to 582 bp (Table 3). The Cyt b fragment of 485 bp showed equal size among all individuals analyzed, and the variable sites were mostly synonymous substitutions at the third codon position (Table 4).
E. townsendi sequences were very different from the other two species, having on average over 20 pairwise differences with E. cordata and E. securigera for both mitochondrial regions, whereas the latter two showed on average less than 11 pairwise differences (Table 5). FST values for the 16S region were significant (P < 0.05) for all pairs of species, demonstrating high interspecific differentiation and therefore supporting the quality of this marker for species identification. The UPGMA distance tree clustered the mitotypes of each species (CA, CB for E. cordata, SM, SN for E. securigera and T for E. townsendi) with high bootstrap values (Fig. 3) indicating low probability of wrong assignment of the species.
For the Cyt b fragment, the average number of interspecific differences (dW) and FST values also showed large differences between E. townsendi and the other two species (Table 5). However, this locus had low interspecific differentiation for E. cordata and E. securigera, as revealed by the non-significant FST value (FST = 0.097; P = 0.17). The UPGMA tree evidenced the strong differences between E. townsendi and the other two species, while E. cordata and E. securigera formed a single group with reticulated mitotypes (Fig. 4).
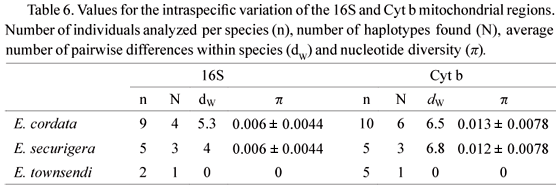
Parameters of intraspecific variation showed that the 16S region was less variable within species than the Cyt b region (Table 6). These estimates validate the use of the 16S region for species identification because, besides the strong differentiation among species, this fragment also showed low intraspecific polymorphism. All of these features make the 16S region an ideal marker for the purpose of molecular identification of species of the genus Euglossa.
Discussion
Identification method. Allozymes proved to be good markers to detect the number of species in our samples, as the absence of heterozygotes in three groups of individuals was an unquestionable evidence of the presence of three distinct species. EST-3 was an important diagnostic marker because each species showed a private fixed allele. Furthermore, ICD 96 allele was diagnostic for E. securigera and MDH 90, ME 94 and PGM 102 alleles were diagnostic for E. townsendi. After characterizing the allozyme patterns for each species, males were used as controls and allowed the simple identification of female individuals.
Variation in sequences coding for the 16S rRNA gene is constrained by its secondary structure but shows high site-to-site and length variation that is absent in protein-coding genes such as Cyt b. For that reason, the 16S gene has been pointed out as a useful marker for phylogenetic analysis among closely related species in Hymenoptera (Whitfield & Cameron 1998). In the present study, the amplified fragment of the 16S region showed diagnostic patterns for the three Euglossa species when digested with VspI, besides showing polymorphism for E. cordata and E. securigera. Therefore, 16S rRNA was an informative locus for species identification.
The Cyt b region did not perform as well as the 16S since E. cordata and E. securigera shared most restriction sites. However, restriction patterns of E. townsendi were very different for all tested endonucleases. These results are probably due to the phylogenetic relationships among the species here analyzed. Although a complete phylogeny of the Euglossa genus is yet to be done, a morphological classification made by Dressler (1978, 1982a, 1982b, 1982c) and Moure (1967) clusters the Euglossa species into six subgenus and 12 species groups. Following that classification, the three species analyzed in this study belong to the subgenus Euglossa s. str. due to the presence of short tongues and the form of the mid-tibial tuff. However, at the species group level, E. townsendi belongs to the purpurea group XI, whereas E. cordata and E. securigera belong to the cordata group XII (Dressler 1982c). The close phylogenetic relation between the latter two species probably explains the resemblance found among their sequences. Dick et al. (2004) obtained similar results after analyzing mitotypes of E. mixta Friese and E. cognata Moure (analis group VIII) for the COI region.
We found diagnostic markers of good quality for the identification of the three Euglossa species floral visitors of T. peruviana. However, before identifying candidate loci for species identification, it should be taken into consideration that DNA markers may have two limitations for this purpose: (1) high intraspecific variation or (2) ancestral polymorphisms. For this study, the 16S region was the best marker for species identification because it showed low intraspecific variation and no shared mitotypes between species. In contrast, the high intraspecific polymorphism and reticulated mitotypes found in the Cyt b region for E. cordata and E. securigera evidenced the retention of ancestral polymorphism, which made impossible the species identification using this locus. Our results suggest that for the Euglossa genus, the cytochrome b gene is informative for identification at the species group level whereas the 16S region is useful at the species level.
The information here provided about the variation in the 16S and cytochrome b sequence should be considered for future DNA barcode analysis since levels of polymorphism of mitochondrial genes differ among bee species. For example, the cytochrome b gene, which was uninformative at the species level in this study, has been successfully used for the identification of Bombus spp. Latreille (Ellis et al. 2006). In the same way, the COI region selected as the standard barcode region (Hebert et al. 2003), although has worked out for identification of bird species (Hebert et al. 2004), may not succeed for the development of euglossine barcodes since this region has shown low divergence rates among closely related species (Dick et al. 2004).
Sampling method. The method here described using T. peruviana flowers as sampling sites of euglossine bees was useful especially for collecting females. Besides the three Euglossa species, Eulaema nigrita Lepeletier, Exaerete smaragdina (Guérin-Méneville) and Eufriesea violacens (Mocsáry) females were also seen visiting T. peruviana flowers in the cities sampled. This type of sampling method may be relevant for research on euglossine bees in urban areas since chemical baits are not strongly attractive to males in open areas such as cities (López-Uribe pers. obs.). Moreover, as it has been noticed by several authors (Ackerman 1983, Roubik & Ackerman 1987, Rebêlo & Garófalo 1997), sampling in flowers is necessary to detect some species that are not attracted to chemical baits. For example, E. townsendi, which is rarely captured using aromatic compounds (Rebêlo & Garófalo 1997), was quite common in the Rifaina samples showing the relevance of plant surveys for species richness evaluation.
The five sampled urban areas showed differences in the number and abundance of euglossine bee species visiting T. peruviana. Nevertheless, this result could be a consequence of different sampling efforts in each city. Nests of E. townsendi have been found in São Carlos and Araras (Del Lama, pers. obs.) suggesting that other species may occur in these cities but were not collected in T. peruviana flowers. Because distribution of bees depends on the availability of resources for food and nest construction, other Euglossa species may have not been sampled because (1) they prefer other nectar sources or (2) T. peruviana trees were not near enough to other food sources they need to survive. These results emphasize the need of data from nests and other food resources to detect species in different ecosystems.
In our urban surveys of T. peruviana plants, we found only three Euglossa species, of which E. cordata was the most common species. The fact that urban areas impose different biotic and abiotic conditions compared to native ecosystems, evidences that the euglossine species that are present in cities tolerate harsh climatic conditions and explore a great variety of food resources. Milet-Pinheiro & Schlindwein (2005) found that E. cordata males left the forest searching for chemical baits in sugarcane monocultures, which indicates that this species easily flies through open areas. However, given that not all species are capable of flying through open areas, the urbanization process probably may have a negative impact on species richness of the euglossine bee community.
The results here presented show a useful genetic tool for the identification of Euglossa spp. at the species level, which is particularly valuable for female individuals. Although our analyses describe patterns only for the species floral visitors of T. peruviana flowers, they can be expanded and applied to other Euglossa species for which morphological identification of females is not currently possible (e.g Euglossa s. str. subgenus). Moreover, our sampling methodology using T. peruviana flowers offers an easy and efficient way to access euglossine females and males for research in urban ecosystems.
Acknowledgments
This paper is part of the M.Sc. thesis project of M. M. López-Uribe at Universidade Federal de São Carlos. We want to thank R. O. Souza and C. A. Oi for their help with collection of samples, J. C. Serrano for taxonomic identification of some specimens, I. C. Godoy and I. F. Lopes for technical help, P. J. Faria for comments on the manuscript and the agencies CNPq (No. 475935/04-7) and CAPES for financial support.
Received 10/VII/06. Accepted 03/V/07.
- Ackerman, J.D. 1983. Diversity and seasonality of male euglossine bees (Hymenoptera: Apidae) in Central Panama. Ecology 64: 274-283.
- Bonilla-Gómez, M.A. & G. Nates-Parra. 1992. Abejas euglosinas de Colombia (Hymenoptera: Apidae) I. Claves ilustradas. Caldasia 17: 149-172.
- Brunner, P.C., C. Fleming & J.E. Frey. 2002. A molecular identification key for economically important thrips species (Thysanoptera: Thripidae) using direct sequencing and a PCR-RFLP-based approach. Agric. For. Entomol. 4: 127-136.
- Cameron, S.A. 2004. Phylogeny and biology of Neotropical orchid bees (Euglossini). Annu. Rev. Entomol. 49: 377-404.
- Crozier, Y.C., S. Koulianos & R.H. Crozier. 1991. An improved test for Africanized honeybee mitochondrial DNA. Experientia 47: 968-969.
- Dick, C.W., D.W. Roubik, K.F. Gruber & A. Bermingham. 2004. Long-distance gene flow and cross-Andean dispersal of lowland rainforest bees (Apidae: Euglossini) revealed by comparative mitochondrial DNA phylogeography. Mol. Ecol. 13: 3375-3785.
- Dowton, M. & A.D. Austin. 1994. Molecular phylogeny of the insect order Hymenoptera: Apocritan relationships. Proc. Natl. Acad. Sci. U.S.A. 91: 9911-9915.
- Dressler, R.L. 1978. An infrageneric classification of Euglossa, with notes on some features of special taxonomic importance (Hymenoptera; Apidae). Rev. Biol. Trop. 26: 187-198.
- Dressler, R.L. 1982a. New species of Euglossa II. (Hymenoptera: Apidae). Rev. Biol. Trop. 30: 121-129.
- Dressler, R.L. 1982b. New species of Euglossa III. The bursigera species group (Hymenoptera: Apidae). Rev. Biol. Trop. 30: 131-140.
- Dressler, R.L. 1982c. New species of Euglossa IV. The cordata and purpurea species groups (Hymenoptera: Apidae). Rev. Biol. Trop. 30: 141-150.
- Ellis, J.S., M.E. Knight, C. Carvell & D. Goulson. 2006. Cryptic species identification: A simple diagnostic tool for discriminating between two problematic bumblebee species. Mol. Ecol. Notes 6: 540-542.
- Gajardo, G., J. Crespo, A. Triantafyllidis, A. Tzika, A. D. Baxevanis, I. Kappas & T.J. Abatzopoulos. 2004. Species identification of Chilean Artemia populations based on mitochondrial DNA RFLP analysis. J. Biogeogr. 31: 547-555.
- Harris, H. & D.A. Hopkinson. 1976. (eds.), Handbook of enzyme electrophoresis in human genetics. North-Holland Publishing, Amsterdam.
- Hebert, P.D.N., M.Y. Stoeckle, T.S. Zemlak & C.M. Francis. 2004. Identification of birds through DNA barcodes. PLoS Biol. 2: 1657-1663.
- Hebert, P.D.N., S. Ratnasingham & J.R. de Waard. 2003. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B. 270 (Suppl.): S96-S99.
- Milet-Pinheiro, P. & C. Schlindwein. 2005. Do euglossine males (Apidae, Euglossini) leave tropical rainforest to collect fragrances in sugarcane monocultures? Rev. Bras. Zool. 22: 853-858.
- Moure, J.S. 1967. Drescrição de algumas espécies de Euglossinae. Atas Simpósio Biota Amazônica 5 (Zool.): 373-394.
- Naeole, C.K. M. & D.S. Haymer. 2003. Use of oligonucleotide arrays for molecular taxonomic studies of closely related species in the oriental fruit fly (Bactrocera dorsalis) complex. Mol. Ecol. Notes 3: 662-665.
- Nei, M. 1987. (eds.), Molecular evolutionary genetics. Columbia University Press, New York, NY, U.S.A, 512p.
- Ramírez, S., R.L. Dressler & M. Ospina. 2002. Abejas euglossinas (Hymenoptera: Apidae) de la región neotropical: Listado de especies con notas sobre su biología. Biota Colombiana 3: 7-118.
- Rebêlo, J.M.M. & C.A. Garófalo. 1997. Comunidades de machos de Euglossini (Hymenoptera: Apidae) em matas semidecíduas do Nordeste de Estado de São Paulo. An. Soc. Entomol. Bras. 26: 243-255.
- Rebêlo, J.M.M. & J.S. Moure. 1995. As espécies de Euglossa Latreille do Nordeste de São Paulo (Apidae, Euglossinae). Rev. Bras. Zool. 12: 445-466.
- Roubik, D.W. & J.D. Ackerman. 1987. Long-term ecology of euglossine orchid-bees (Apidae:Euglossini) in Panama. Oecologica 73: 321-333.
- Roubik, D.W. & P.E. Hanson. 2004. (eds.), Orchid bees of tropical America. Biology and field guide. Instituto Nacional de Biodiversidade (INBio), Santo Domingo de Heredia, Costa Rica, 370p.
- Schama, R., A.M. Solé-Cava & J.P. Thorpe. 2005. Genetic divergence between east and west Atlantic populations of Actinia spp. sea anemones (Cnidaria: Actiniidae). Mar. Biol. 146: 435-443.
- Schneider, S., D. Roessli & L. Excoffier. 2000. Arlequin ver. 2.000: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- Sheppard, W.S. & B.A. McPheron. 1991. Ribosomal DNA diversity in Apidae, p.89-102. In D.R. Smith (eds.), Diversity in the genus Apis Westview, Boulder, 265p.
- Sites, J.W. & J.C. Marshall. 2003. Delimiting species: a Renaissance issue in systematic biology. Trends Ecol. Evol. 18: 462-470.
- Swofford, D.L. 1999. PAUP*: Phylogenetic Analysis Using Parsimony 4.0b (*and Other Methods). Sinauer Associates, Sunderland, MA.
- Thomson, J.D., T.J. Gibons, F. Plewniak, F. Jeanmougin & D.G. Higgins. 1997. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876-4882.
- Weir, B.S. & C.C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:995-1005.
- Whitfield, J.B. & S.A. Cameron. 1998. Hierarchical analysis of variation in the mitochondrial 16S rRNA gene among Hymenoptera. Mol. Biol. Evol. 15: 1728-1743.
- Zayed, A., D.W. Roubik & L. Packer. 2004. Use of diploid male frequency data as an indicator of pollinator decline. Proc. R. Soc. Lond. B. 271(Suppl.): S9-S12.
Publication Dates
-
Publication in this collection
29 Nov 2007 -
Date of issue
Oct 2007
History
-
Accepted
03 May 2007 -
Received
10 July 2006