Abstracts
We have examined phylogenetic relationships in seven pathogenesis-related (PR) protein families. Within-family comparisons involved 79 species, 166 amino acid sequences, and 1,791 sites. For 37 species, 124 different PR isoforms were identified (an average of 3.3 per species). Thirty-one of the 37 species investigated tended to cluster together (84%). Of the 17 clusters distinguished in the seven phylogenetic trees, 10 (59%) were in agreement with their taxonomic status, ascertained at the family level. The strong similarities among the intraspecific forms, as compared to interspecific differences, argue for some kind of gene conversion, but the rare occurrence of widely different isoforms also suggests diversifying selection. PRs 1, 6, and 4 seem to be less differentiated than PRs 3, 2, 10, and 5.
pathogenesis-related proteins; PR; molecular evolution; plant defense
Foram analisadas as relações filogenéticas em sete famílias de proteínas relacionadas à patogênese. As comparações dentro das famílias envolveram 79 espécies, 166 seqüências de aminoácidos e 1.791 sítios nucleotídicos. Para 37 espécies, foram identificadas 124 isoformas diferentes de PRs (uma média de 3,3 por espécie). Trinta e uma (84%) das investigadas nas 37 espécies tenderam a se agrupar. Dos 17 agrupamentos diferenciados nas sete árvores filogenéticas, 10 (59%) estiveram de acordo com a classificação taxonômica, avaliada em nível de família. A forte similaridade entre as formas intraespecíficas, quando comparadas às diferenças interespecíficas, sugere algum tipo de conversão gênica, mas a ocorrência rara de isoformas muito diferentes pode também sugerir seleção diversificadora. As PRs 1, 6 e 4 parecem ser menos diferenciadas do que as PRs 3, 2, 10 e 5.
proteínas relacionadas à patogênese; PRs; evolução molecular; defesa em plantas
Evolutionary implications of infra- and interspecific molecular variability of pathogenesis-related proteins
Implicações evolutivas da variabilidade molecular intra e interespecífica de proteínas relacionadas à patogênese
Freitas, L. B.I; Bonatto, S. L.II; Salzano, F. M.I
IDepartamento de Genética, Instituto de Biociências, Universidade Federal do Rio Grande do Sul, C.P. 15053, CEP 91501-970, Porto Alegre, RS, Brazil
IICentro de Biologia Genômica e Molecular, Faculdade de Biociências, Pontifícia Universidade Católica do Rio Grande do Sul, Av. Ipiranga, 6681, CEP 90610-001, Porto Alegre, RS, Brazil
Correspondence Correspondence to Loreta B. Freitas Departamento de Genética, UFRGS, C.P. 15053 CEP 91501-970, Porto Alegre, RS, Brazil e-mail: loreta.freitas@ufrgs.br
ABSTRACT
We have examined phylogenetic relationships in seven pathogenesis-related (PR) protein families. Within-family comparisons involved 79 species, 166 amino acid sequences, and 1,791 sites. For 37 species, 124 different PR isoforms were identified (an average of 3.3 per species). Thirty-one of the 37 species investigated tended to cluster together (84%). Of the 17 clusters distinguished in the seven phylogenetic trees, 10 (59%) were in agreement with their taxonomic status, ascertained at the family level. The strong similarities among the intraspecific forms, as compared to interspecific differences, argue for some kind of gene conversion, but the rare occurrence of widely different isoforms also suggests diversifying selection. PRs 1, 6, and 4 seem to be less differentiated than PRs 3, 2, 10, and 5.
Key words: pathogenesis-related proteins, PR, molecular evolution, plant defense.
RESUMO
Foram analisadas as relações filogenéticas em sete famílias de proteínas relacionadas à patogênese. As comparações dentro das famílias envolveram 79 espécies, 166 seqüências de aminoácidos e 1.791 sítios nucleotídicos. Para 37 espécies, foram identificadas 124 isoformas diferentes de PRs (uma média de 3,3 por espécie). Trinta e uma (84%) das investigadas nas 37 espécies tenderam a se agrupar. Dos 17 agrupamentos diferenciados nas sete árvores filogenéticas, 10 (59%) estiveram de acordo com a classificação taxonômica, avaliada em nível de família. A forte similaridade entre as formas intraespecíficas, quando comparadas às diferenças interespecíficas, sugere algum tipo de conversão gênica, mas a ocorrência rara de isoformas muito diferentes pode também sugerir seleção diversificadora. As PRs 1, 6 e 4 parecem ser menos diferenciadas do que as PRs 3, 2, 10 e 5.
Palavras-chave: proteínas relacionadas à patogênese, PRs, evolução molecular, defesa em plantas.
INTRODUCTION
Plants have developed natural defense mechanisms acting at multiple levels in the prevention of pathogen colonization and disease. These mechanisms involve passive and active, constitutive, and inducible elements. Defense genes can be subdivided in three classes: (a) those whose product directly changes the cell matrix properties, therefore influencing the physical barriers imposed by the cell; (b) genes which codify proteins with antimicrobial activity or catalyze the synthesis of antibiotic products; they include enzyme inhibitors (amylases, proteinase inhibitors), toxic proteins (lectins, thionins), hydrolases (chitinases, glucanases), proteinases, and enzymes influencing the biosynthesis of oxy-phenols, tannins, ortho-chinones, and phytoalexins; and (c) those that codify pathogenesis-related (PR) proteins (Baron & Zambryski, 1995; Kombrink & Somssich, 1995; Swords et al., 1997). Besides the defense genes, about 20 resistance genes have been described and analyzed. Their molecular characterizations have indicated many similarities between them, and the study of their evolutionary patterns has suggested diversifying selection, unequal crossing-over, and gene conversion as the main mechanisms which generate diversity (Michelmore & Meyers, 1998).
Pathogenesis-related proteins were first detected in the early 1970s in tobacco leaves reacting hypersensitively to tobacco mosaic virus; these were variously named until Antoniw et al. (1980) proposed the term used until now. The PR proteins display characteristic physico-chemical properties which help in their detection and isolation. They are: (a) very stable at low (around 3.0) pHs; (b) relatively resistant to endogenous and exogenous proteolytic enzyme action; (c) generally monomers of low molecular mass (8-100 kDa); and (d) preferentially localized in intercellular spaces. While these proteins are not generally detected in healthy plants, in those infected or submitted to specific chemical treatment they may account for 10% of the leaf content in soluble proteins (Stintzi et al., 1993).
At the 3rd International Workshop on PR proteins (Arolla, Switzerland, August 16-20, 1992) it was agreed that common classification and nomenclature should be used. Specifically, five families were recognized, based on different criteria. The numbering was according to relative mobility in nondenaturing gel systems. Those classified in a given family are serologically related, have very similar molecular weights, and share highly uniform amino acid sequences (Linthorst, 1991; Cutt & Klessig, 1992).
With investigative progress, new proteins have been identified, and at present 14 PR families are officially recognized (Van Loon & Van Strien, 1999), although new proteins with potential anti-pathogenic action continue to be described at a rapid pace.
Increased interest in this biological system arose as it was verified that PRs are not restricted to plants, but are also present in fungi and animals (invertebrate and vertebrate), constituting the so-called ''PR protein superfamily''. Its members include proteins from fungi (Saccharomyces, Schizophylum) and nematodes (Caenorhabditis); antigen 5, one major vespid venom allergen and an antigen 5-related protein from Drosophila melanogaster; helothermine, from a lizard venom; mammalian Tpx-1 testis-specific protein and sperm-coating glycoprotein Scg; and human specific granule protein 28 from neutrophils, P25TI trypsin inhibitor of neuroblastoma and glioblastoma cells, and glioma pathogenesis-related GliPr (Schreiber et al., 1997; Szyperski et al., 1998; Yamakawa et al., 1998).
Considering their ubiquity and importance, it is surprising to verify that no systematic molecular comparison of these proteins has been undertaken, the notable exception being the thorough analysis performed by Wen et al. (1997) on the Bet v 1 homologues (PR 10), and by Bishop et al. (2000) on the chitinases of PR8. Here we consider the molecular relationships of PR10 and six other PR families.
MATERIALS AND METHODS
The amino acid sequences considered were obtained from the Prosite data bank (http://expasy.hcuge.ch/sprot/prositc.html) which groups all protein sequences described in Swiss-Prot (http://expasy.hcuge.ch/sprot/sprot-top.html), in accordance with their signatures (characteristic sequences of each protein group). The alignment was made using the ClustalW program, and Kimura's (1983) genetic distance was determined among the sequences. Phylogenetic trees were obtained by the neighbor-joining method (Saitou & Nei, 1987). Matrices of genetic distances and the phylogenies were constructed using the Treecon for Windows program.
RESULTS
The proteins considered, classified in seven families, are listed in Tables 1-7; the corresponding dendrograms are shown in Figs. 1-7. Each PR family will be considered separately, and at the end we will determine what generalizations can be made.
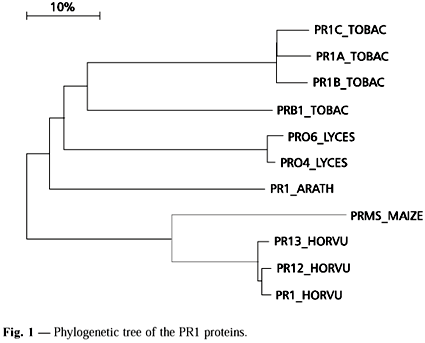
Eleven PR1 sequences comprising 172 sites each were evaluated. They occur in five species, distributed among three taxonomic families. Three species present a total of nine isoforms, all of them occurring together in the dendrogram. The two clusters observed are formed by proteins from species belonging to the same taxonomic family (upper, Solanaceae; lower, Gramineae). The PR1 from Arabidopsis thaliana cluster with those from the Solanaceae.
For PR2, 35 sequences, and 344 sites were compared (Table 2 and Fig. 2). They are distributed along 14 species, classified in six families. Seven species present 28 multiple PR forms; of these, the isoforms of Nicotiana plumbaginifolia (NICPL), Solanum tuberosum (SOLTU), and Hordeum sativum (HORVU) occur together. Those from Nicotiana tabacum (TOBAC) form two main groups, but those from Glycine max (SOYBN), Arabidopsis thaliana (ARATH), and Solanum lycopersicum (LYCES) occur separately. Of the three clusters formed, one includes two subclusters of Solanaceae, but the third subcluster is mixed (shows proteins from different taxonomic families).
The second cluster is formed basically by Gramineae proteins, while the third is mixed (ARATH and WHEAT, the latter from Triticum aestivum).
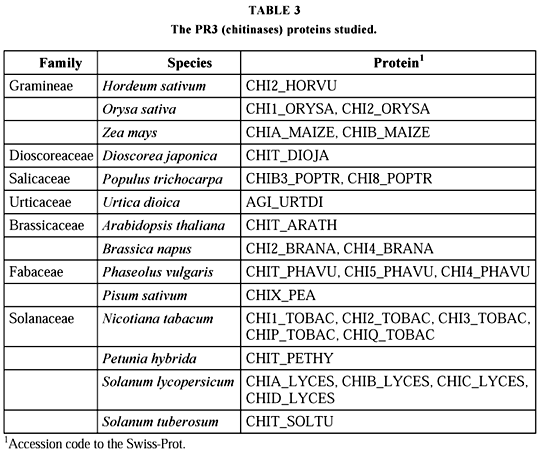
A total of 27 PR3 sequences comprising 391 sites could be assembled (Table 3 and Fig. 3). They are present in 14 species, grouped in seven families. Seven of the species present 20 isoforms, and of these ORYSA (from Orysa sativa) and MAIZE (Zea mays) occur together. TOBAC, LYCES, and PHAVU (the latter from Phaseolus vulgaris) are each present in two different clusters, while BRANA (from Brassica napus), and POPTR (Populus trichocarpa) appear in different clusters. Two main clusters are visualized, the first (upper part of the figure), larger, subdivided in four subclusters. The first and fourth subclusters are composed by Solanaceae and the third by Gramineae proteins. The second is mixed (BRANA + ARATH + PHAVU). The other cluster is also mixed (DIOJA, from Dioscorea japonica, + PHAVU + BRANA + MAIZE).
Less information is available for PR4. The results include nine sequences (217 sites compared), distributed among seven species belonging to five families (Table 4 and Fig. 4). Two species show four isoforms which occur together. Little differentiation was observed, with the Hordeum (HORVU) and Glycine (SOYBN) proteins being set somewhat apart.
A total of 26 PR5 sequences comprising 250 sites, in 12 species of six families, were compared (Table 5 and Fig. 5). The 22 multiple forms from the same species (eight considered) generally occur together, with those from Arabidopsis (ARATH) and Nicotiana (TOBAC) clustering in two groups. Two main clusters can be observed, the first (upper part of the figure) presenting two differentiated subclusters. The one at the top is composed mainly by Solanaceae, while the second unites ARATH with PRUAV (from Prunus avium, classified in a different taxonomic family). The second cluster is composed by Gramineae proteins.
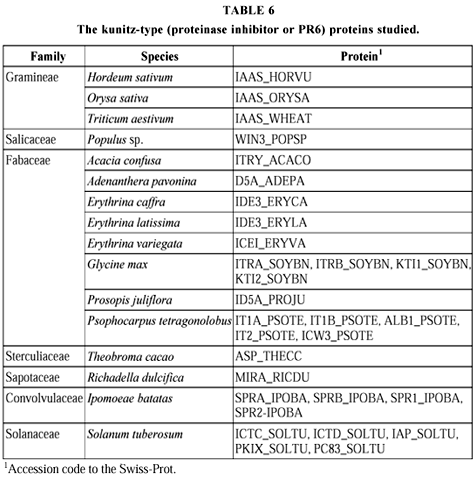
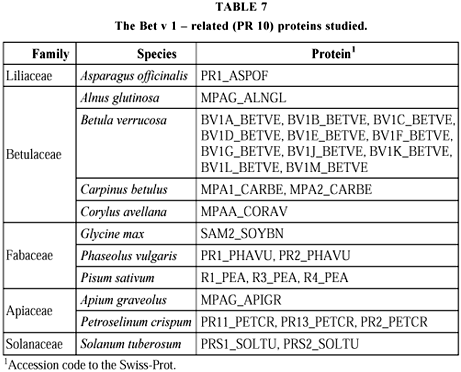
Thirty PR6 sequences with 255 sites, found in 16 species classified in seven families were compared (Table 6 and Fig. 6). The 18 multiple forms from the four species in which they occur are generally found together (SOYBN, SOLTU, from Solanum tuberosum, IPOBA, from Ipomoea batatas), while those from Psophocarpus tetragonolobus are distributed in two groups of two, with another (ALB1_PSOTE) quite separated. Three clusters can be distinguished. The first is composed by Fabaceae proteins, the second by Solanum tuberosum forms, while in the third a subcluster is formed by Gramineae proteins; the others show proteins of species from different families. For PR10 there is complete congruence with the taxonomic classification (Table 7 and Fig. 7). Twenty-eight sequences with 162 sites, observed in 11 species which belong to five families, were studied. Six species presented 23 isoforms, and all clustered together, especially BTVE, which showed little differentiation. In the three clusters formed, the first and third are respectively comprised by Betulaceae and Fabaceae proteins. In the second, one subcluster is formed by Apiaceae and the other by Solanaceae proteins, ASPOF, from Asparagus officinalis, forming a separate branch.
General overview
We, therefore, examined seven PR families. Within-family comparisons involved 79 species, 166 sequences, and 1,791 sites. For 37 species, 124 different PR isoforms were identified (an average of 3.3 per species). Thirty-one of those investigated in the 37 species tended to cluster together (84%). Of the 17 clusters distinguished in the seven phylogenetic trees, 10 (59%) were clearly in agreement with the taxonomy as evaluated at the family level.
What comparisons can be made among the PR families? The average genetic distances obtained clearly separates them into two groups (despite the expected high standard deviations) apparently unrelated to their functions: group 1: PR1 (function unknown), 0.02 ± 0.02; PR 6 (proteinase inhibitors), 0.10 ± 0.03; and PR4 (chitinases), 0.26 ± 0.09; group 2: PR3 (chitinases), 0.73 ± 0.32; PR2 (b-1,3-glucanases), 0.76 ± 0.32; PR10 (birch allergen Bet v 1-related), 0.77 ± 0.43; and PR5 (thaumatin-like), 0.98 ± 0.50.
DISCUSSION
The genetic system underlying the PR proteins clearly qualifies for classification as a multigene family, since there is a multiplicity of forms which occur both within and between species, with a general molecular similarity among them. The mechanisms that may give origin to such families have been discussed by Li (1997); these include saltatory replication, unequal crossing-over, replication slippage, gene conversion, and duplicative transposition. Without direct evidence it is impossible, at the moment, to indicate any one of them as responsible for the PR system.
The degree of variability found, however, allows some inferences about the evolution of these forms. Thus, the strong similarities among the intraspecific isoforms (84% of them clustered together in the phylogenetic trees) as compared to the interspecific comparisons (only about half of the cases were congruent with their taxonomic status as assessed at the family level) suggest a process of gene conversion (Zimmer et al., 1980).
Equally interesting, however, are the cases in which the isoforms did not cluster together. Marked differences, for instance, were observed between the PR2 isoforms of Arabidopsis thaliana ARATH_E132 and ARATH_EA6 (genetic distance of 1.16), and the PR3 isoforms of Brassica napus BRANA_CHI2 and BRANA_CHI4 (genetic distance of 1.06). Less pronounced but still important, are the differences between the PR2 isoforms of Glycine max E13A_SOYBN and E13B_SOYBN (genetic distance of 0.82), and the PR3 isoforms of Populus trichocarpa, CHIB_POPTR and CHI8_POPTR (genetic distance of 0.75). Due to the special characteristics of this genetic system, the occurrence of diversifying selection between these forms does not seem unreasonable. We have considered this question in detail in another study (Scherer et al., 2003).
Our results also suggest that PRs 1, 6, and 4 are less differentiated than PRs 3, 2, 10, and 5. In terms of differentiating mechanisms, Michelmore & Meyers' (1998) model for resistance genes may well also apply to PR genes. It proposes initial diversification through interallelic recombination and gene conversion, followed by divergent evolution of individual genes and a birth-and-death process.
Some of the PRs considered here have been phylogenetically analysed by other authors. The extended analysis of 67 PR10 sequences performed by Wen et al. (1997) at the nucleotide level has already been mentioned. Hoffmann-Sommergruber et al. (1997) also considered seven amino acid sequences of this same family, from an equivalent number of species.
The present analysis generally agrees with the relationships those authors obtained. On the other hand, the study of Szyperski et al. (1998) was mainly concerned with a global genealogical tree of the PR protein superfamily, which includes the plant PR1. They considered five protein sequences of the latter in an equivalent number of species and obtained phylogenetic relationships that have basically been confirmed by us.
It is not yet clear how the PR protein superfamily evolved. But Szyperski et al. (1998) have found that human GliPR and plant PR1 proteins seem to operate according to the same molecular mechanism, suggesting a functional link between the human immune and plant defense systems. Convergent evolution of two independent ancestors is unlikely. Therefore, the other possibilities are that either P14a (the PR1 protein studied) and GliPR arose from a common ancestor at a very early stage of evolution, or that horizontal transfer occurred at a much later stage.
At present it is impossible to decide between these two hypotheses. But the evolutionary importance of transposons is increasingly being recognized, as exemplified by the studies of Clegg et al. (1997) and Grandbastien (1998).
As was stressed by Stintzi et al. (1993) and Swords et al. (1997), understanding how plants resist outside insults may open the way for genetically-engineered species with improved resistance against fungi and bacteria, obtained by transformation with PR genes. The aim could be PR combinations that would either act synergistically, or differentially attack microbes. To achieve this goal, it is important to know the type of variability present within and between species, as well as the differences that exist between different family sets. The present study is a modest contribution in that direction.
Acknowledgments This research was funded by the Programa de Apoio a Núcleos de Excelência (PRONEX), Financiadora de Estudos e Projetos (FINEP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), and Pró-Reitoria de Pesquisa, Universidade Federal do Rio Grande do Sul (PROPESQ-UFRGS).
SCHERER, N. M., FREITAS, L. B., SALZANO, F. M. & BONATTO, S. L., 2003, Patterns of molecular evolution in pathogenesis-related proteins (submitted for publication).
Received April 1, 2002
Accepted May 20, 2002
Distributed August 31, 2003
- ANTONIW, J. F., RITTER, C. E., PIERPOINT, W. S. & VAN LOON, L. C., 1980, Comparison of three pathogenesis-related proteins from plants of two cultivars of tobacco infected with TMV. J. Gen. Virol., 47: 79-87.
- BARON, C. & ZAMBRYSKI, P. C., 1995, The plant response in pathogenesis, symbiosis, and wounding: Variations on a common theme? Annu. Rev. Genet., 29: 107-129.
- BISHOP, J. G., DEAN, A. M. & MITCHELL-OLDS, T., 2000, Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA, 97: 5322-5327.
- CLEGG, M. T., CUMMINGS, M. P. & DURBIN, M. L., 1997, The evolution of nuclear genes. Proc. Natl. Acad. Sci. USA, 94: 7791-7798.
- CUTT, J. R. & KLESSIG, D. F., 1992, Pathogenesis-related proteins, pp. 209-243. In: T. Boller & F. Meins (eds.), Genes Involved in Plant Defense Springer-Verlag, Berlin.
- GRANDBASTIEN, M. A., 1998, Activation of plant retrotransposons under stress conditions. Tr. Plant Sci., 3: 181-187.
- HOFFMANN-SOMMERGRUBER, VANEK-KREBITZ, M., RADAUER, C., WEN, J., FERREIRA, F., SCHEINER, O. & BREITENEDER, H., 1997, Genomic characterization of the Bet v 1 family: Genes coding for allergens and pathogenesis-related proteins share intron positions. Gene, 197: 91-100.
- KIMURA, M., 1983, The neutral theory of molecular evolution Cambridge University Press, Cambridge, 328p.
- KOMBRINK, E. & SOMSSICH, I. E., 1995, Defense responses of plants to pathogens. Adv. Bot. Res., 21: 1-34.
- LI, W. H., 1997, Molecular evolution Sinauer Associates, Sunderland, Massachusetts, 487p.
- LINTHORST, H. J. M., 1991, Pathogenesis-related proteins of plants. Crit. Rev. Plant Sci., 10: 123-150.
- MICHELMORE, R. W. & MEYERS, B. C., 1998, Clusters of resistance genes in plants evolve by divergent selection and birth-and-death process. Gen. Res., 8: 1113-1130.
- SAITOU, N. & NEI, M., 1987, The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4: 406-425.
- SCHREIBER, M. C., KARLO, J. C. & KOVALICK, G. E., 1997. A novel cDNA from Drosophila encoding a protein with similarity to mammalian cysteine-rich secretory proteins, wasp venom antigen 5, and plant group 1 pathogenesis-related proteins. Gene, 191: 135-141.
- STINTZI, A., HEITZ, T., PRASAD, V., WIEDEMANN-MERDINOGLU, S., KAUFFMANN, S., GEOFFROY, P., LEGRAND, M. & FRITIG, B., 1993, Plant ''pathogenesis-related'' proteins and their role in defense against pathogens. Biochimie, 75: 687-706.
- SWORDS, K. M. M., LIANG, J. & SHAH, D. M., 1997, Novel approaches to engineering disease resistance in crops. Genet. Engin., 19: 1-13.
- SZYPERSKI, T., FERNÁNDEZ, C., MUMENTHALER, C. & WÜTHRICH, K., 1998, Structure comparison of human glioma pathogenesis-related protein GliPR and the plant pathogenesis-related protein P14a indicates a functional link between the human immune system and a plant defense system. Proc. Natl. Acad. Sci., USA, 95: 2262-2266.
- VAN LOON, L. C. & VAN STRIEN, E. A., 1999, The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol., 55: 85-97.
- WEN, J., VANEK-KREBITZ, M., HOFFMANN-SOMMERGRUBER, K., SCHEINER, O. & BREITENEDER, H., 1997, The potential of Bet v 1 homologues, a nuclear multigene family, as phylogenetic markers in flowering plants. Molec. Phylogenet. Evol., 8: 317-333.
- YAMAKAWA, T., MIYATA, S., OGAWA, N., KOSHIKAWA, N., YASUMITSU, H., KANAMORI, T. & MIYAZAKI, K., 1998, cDNA cloning of a novel trypsin inhibitor with similarity to pathogenesis-related proteins, and its frequent expression in human brain cancer cells. Bioch. Biophys. Acta, 1395: 202-208.
- ZIMMER, E. A., MARTIN, S. L., BEVERLY, S. M., KAN, Y. W. & WILSON, A. C., 1980, Rapid duplication and loss of genes coding for the a chains of hemoglobin. Proc. Natl. Acad. Sci. USA, 77: 2158-2162.
Publication Dates
-
Publication in this collection
20 Jan 2004 -
Date of issue
Aug 2003
History
-
Accepted
20 May 2002 -
Received
01 Apr 2002