Abstracts
Cytogenetic analyses of the location of 18S and 5S ribosomal DNAs, and the base composition of B chromosomes of Iheringichthys labrosus from Tibagi River, Paraná, Brazil, are provided. AgNORs were observed in the terminal position on the long arm of a subtelocentric chromosome pair. CMA3-positive staining was observed in some chromosomes, which besides being associated with NORs, were all DAPI-negative. Chromosome B showed a strong fluorescence with CMA3. The concomitant use of 18S and 5S rDNA probes using the FISH technique revealed 18S ribosomal cistrons in a pair of subtelocentric chromosomes, on the long arm in the terminal position, coinciding with the AgNOR. The 5S sites were found in another subtelocentric pair, on the long arm in the interstitial region, near the centromere. The findings of the present study suggest that, although there are some more conserved cytogenetic characteristics, populations of I. labrosus may show their own characteristics.
supernumerary chromosome; catfish; FISH
Foram realizadas análises citogenéticas em Iheringichthys labrosus do Rio Tibagi, Paraná, Brasil com a localização cromossômica dos DNAs ribossômicos 18S e 5S e a composição de bases de seus cromossomos B. As AgNORs foram observadas em posição terminal, no braço longo de um par de cromossomos subtelocêntricos. Marcações CMA3 positivas foram observadas em alguns cromossomos e associadas com as RONs. Porém, todas estas marcações apresentaram-se DAPI negativas. O cromossomo B mostrou-se fortemente fluorescente com CMA3. O uso concomitante das sondas de DNAr 18S e 5S, através da técnica de FISH, revelou os cístrons ribossômicos em um par de cromossomos subtelocêntricos, em posição terminal do braço longo, coincidindo com a AgNOR. Os sítios 5S foram observados em outro par subtelocêntrico, em posição intersticial do braço longo, próximo ao centrômero. Os resultados observados no presente estudo sugerem que, embora existam algumas características citogenéticas mais conservadas, as populações de I. labrosus podem mostrar suas próprias características.
cromossomo supranumerário; catfish; FISH
BIOLOGY
Cytogenetic analysis of the 18S, 5S rDNA and B chromosome of Iheringichthys labrosus (Lütken, 1874) (Siluriformes, Pimelodidae)
Análise Citogenética de DNAr 18S, 5S e cromossomo B de Iheringichthys labrosus (Lütken, 1874) (Siluriformes, Pimelodidae)
Carvalho, RA.I, II; Laudicina, A.III; Giuliano-Caetano, L.II; Martins-Santos, IC.I; Dias, AL.II, * * e-mail: anadias@uel.br
IDepartamento de Biologia Celular e Genética, Centro de Ciências Biológicas - CCB, Universidade Estadual de Maringá - UEM, Av. Colombo, 5790, CEP 87020-900, Maringá, PR, Brazil
IIDepartamento de Biologia Geral, Universidade Estadual de Londrina - UEL, Campus Universitário. Rodovia Celso Garcia Cid, Km 380, CEP 86055-900, Londrina, PR, Brazil
IIIDepartamento de Radiologia, Comisión Nacional de Energía Atômica, San Martín, Buenos Aires, Argentina
ABSTRACT
Cytogenetic analyses of the location of 18S and 5S ribosomal DNAs, and the base composition of B chromosomes of Iheringichthys labrosus from Tibagi River, Paraná, Brazil, are provided. AgNORs were observed in the terminal position on the long arm of a subtelocentric chromosome pair. CMA3-positive staining was observed in some chromosomes, which besides being associated with NORs, were all DAPI-negative. Chromosome B showed a strong fluorescence with CMA3. The concomitant use of 18S and 5S rDNA probes using the FISH technique revealed 18S ribosomal cistrons in a pair of subtelocentric chromosomes, on the long arm in the terminal position, coinciding with the AgNOR. The 5S sites were found in another subtelocentric pair, on the long arm in the interstitial region, near the centromere. The findings of the present study suggest that, although there are some more conserved cytogenetic characteristics, populations of I. labrosus may show their own characteristics.
Keywords: supernumerary chromosome, catfish, FISH.
RESUMO
Foram realizadas análises citogenéticas em Iheringichthys labrosus do Rio Tibagi, Paraná, Brasil com a localização cromossômica dos DNAs ribossômicos 18S e 5S e a composição de bases de seus cromossomos B. As AgNORs foram observadas em posição terminal, no braço longo de um par de cromossomos subtelocêntricos. Marcações CMA3 positivas foram observadas em alguns cromossomos e associadas com as RONs. Porém, todas estas marcações apresentaram-se DAPI negativas. O cromossomo B mostrou-se fortemente fluorescente com CMA3. O uso concomitante das sondas de DNAr 18S e 5S, através da técnica de FISH, revelou os cístrons ribossômicos em um par de cromossomos subtelocêntricos, em posição terminal do braço longo, coincidindo com a AgNOR. Os sítios 5S foram observados em outro par subtelocêntrico, em posição intersticial do braço longo, próximo ao centrômero. Os resultados observados no presente estudo sugerem que, embora existam algumas características citogenéticas mais conservadas, as populações de I. labrosus podem mostrar suas próprias características.
Palavras-chave: cromossomo supranumerário, catfish, FISH.
1. Introduction
The genus Iheringichthys belongs to the order Siluriformes, family Pimelodidae, and includes only two species: Iheringichthys megalops Eigenmann and Ward, 1907, which is found in the Paraguay River basin, and Iheringichthys labrosus, in the Paraná River basin (Burgess, 1989). Only some populations of I. labrosus have been studied from a cytogenetic perspective, demonstrating a diploid number of 56 chromosomes, with the presence of up to three supernumerary chromosomes or Bs (Vissotto et al., 1999; Carvalho et al., 2004; Carvalho and Dias, 2005a).
The supernumerary chromosomes or Bs have been described in 61 species of Neotropical freshwater fishes (Carvalho et al., 2008). Only five species among those of the family Pimelodidae have such chromosomes: Bergiaria westermanni (Lütken, 1874) (Dias and Foresti, 1993), Iheringichthys labrosus (Vissotto et al., 1999; Carvalho et al., 2004; Carvalho and Dias, 2005a), Pimelodus ortmanni Haseman, 1911 and Pimelodus sp. NUP690, NUP1664, NUP1786, NUP1826 (NUP refers to the ichthyological collection of NUPELIA, Núcleo de Pesquisas em Limnologia, Ictiologia e Aquicultura of Universidade Estadual de Maringá, PR, Brazil) (Borin and Martins-Santos, 2004) and Megalonema platanum (Günther, 1880) (Carvalho, 2008). There are few papers that have examined the base pair composition of these chromosomes in fish (Vênere et al., 1999; Maistro et al., 2000, 2002; Jesus et al., 2003).
Fluorochromes have been used to demonstrate chromosomal regions rich in certain pairs of nitrogenous bases, since they stain preferentially specific regions. In fish, staining with GC-specific fluorochromes, such as CMA3, has shown a close relationship between the nucleolar organizer regions (NORs) observed by impregnation with silver nitrate (AgNO3) and the chromosomal regions stained with these fluorochromes. In some species of the family Pimelodidae, fluorochromes have also revealed several other regions, which appear to be related to a specific type of heterochromatin (Swarça et al., 2003; Souza et al., 2004; Treco et al., 2008).
In higher eukaryotes, tandem arrays of ribosomal RNA genes are organised into two different multigene families composed of hundreds of thousands of copies. One family is represented by 45S rDNA encoding 18S, 5.8S and 26S/28S rRNAs, and the other is represented by 5S rDNA that encodes 5S rRNA, one of the largest components of ribosome subunits. The 5S rDNA arrangement consists of sequences of 120 base pairs (bp) that are separated from each other by non-transcribed spacer regions (Martins and Galetti-Júnior, 2001).
Although NORs are generally viewed by silver impregnation and by fluorochromes that stain preferably GC-rich regions, they can also be located through the technical accuracy of fluorescent in situ hybridisation (FISH) with 45S, 28S and 18S rDNA probes. The ribosomal cistrons of 5S rDNA can only be identified using the FISH technique, and are therefore not related to NORs.
The objective of this study was to examine the chromosomal location of 18S and 5S ribosomal DNAs, and the base composition of B chromosomes in a population of Iheringichthys labrosus from the Tibagi River basin (Paraná, Brazil), and compare the results with those obtained for the population of the Capivara Reservoir, previously reviewed by Carvalho and Dias (2005a, b, 2007).
2. Material and Methods
Eight individuals from Iheringichthys labrosus, collected in the Tibagi River, Londrina, Paraná, Brazil (23° 21' 27.22'' S and 51° 00' 34. 36'' W) were analysed. Mitotic chromosomes were obtained from kidney cells according to the conventional technique of Bertollo et al. (1978). Nucleolar organizer regions (NORs) were identified by silver nitrate staining according to Howell and Black (1980). Fluorochrome staining with the GC- specific chromomycin - A3 (CMA3) and AT-specific diamidino-2-phenylindole (DAPI) was carried out according to Schweizer (1980). The 18S rDNA (1700 bp) of Oreochromis niloticus niloticus (Linnaeus, 1758) and 5S rDNA (200 bp) of Leporinus elongates Valenciennes, 1850, were used concurrently for the fluorescent in situ hybridisation, following the protocol described by Christian et al. (1998).
3. Results
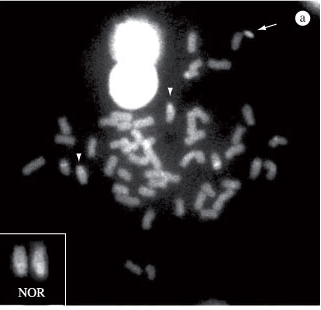
The AgNOR in I. labrosus from the Tibagi River was observed on the long arm of a pair of subtelocentric chromosomes, in the terminal position (Figure 1a). The concomitant use of 18S and 5S rDNA probes, using the FISH technique, showed the 18S ribosomal cistrons in a pair of subtelocentric chromosomes, on the long arm in the terminal position (Figure 1b) and the 5S sites were found in another subtelocentric pair, on the long arm in the interstitial region, near the centromere (Figure 1b).
Some chromosomes showed CMA3 positive sites after fluorochrome treatment, mainly at one terminal and a few interstitial positions. A pair of subtelocentric chromosomes showed terminal staining on the long arm (Figure. 2a). With the fluorochrome DAPI, this same pair showed negative staining (Figure. 2b). The analysis of metaphases with a single B chromosome in the population from the Tibagi River revealed that this chromosome is strongly fluorescent with CMA3 and negative with DAPI (Figures 2a, b).
4. Discussion
Carvalho et al. (2004) has previously described the karyotype of I. labrosus from the Tibagi River, which showed a diploid number of 2n = 56 chromosomes with karyotype formula of 32 m + 8 sm + 6st + 10a, and also described the presence of 0-3 supernumerary microchromosomes in this population. The same diploid number was found in all populations of this examined species, but different karyotype formulas were observed (Table 1). Such differences may be due to processes of chromosomal rearrangements that do not change the diploid number, such as pericentric inversions and/or translocations. Different karyotype formulas may also suggest that this is a species complex that requires a taxonomic revision in Iheringichthys labrosus.
Table 1 shows that supernumerary chromosomes occur in 75% of the populations of I. labrosus. Among these populations carrying B chromosomes, 70% of all individuals examined had such chromosomes, representing a high interindividual frequency of B chromosomes in this species.
The AgNOR has revealed to be conserved regarding its location on the long arm of a pair of subtelocentric chromosomes, as corroborated by the present study, in the same way as other populations of I. labrosus previously analysed (Table 1). One possible exception is the population from the Guaraúna River, Paraná, where the NOR is found on the short arm of a submetacentric pair (Ribeiro et al., 2008). In the family Pimelodidae, the position of the NOR seems to represent an important factor in distinguishing between species of large size, such as the group "Sorubiminae" that, without exception, has the NOR on the short arms, and species of small to medium size, such as the group "Pimelodus", which fits in the genus Iheringichthys and most usually has the NOR on the long arm (Fenocchio et al., 2003).
In the I. labrosus population of the Capivara Reservoir reviewed by Carvalho and Dias (2007), hybridisation with the 18S rDNA probe also showed signals corresponding to AgNORs in a pair of st-chromosomes and on the long arm in the terminal position, as in the population from the Tibagi River. The 5S rDNA location was different from that of the population studied herein, because the 5S ribosomal cistrons were in a terminal position on the long arm of a subtelocentric pair.
The use of the 18S rDNA probe has been growing in the family Pimelodidae, confirming data obtained with silver nitrate, as observed in Pinirampus pirinampu (Spix and Agassiz, 1829) (Swarça et al., 1999), Zungaro zungaro (Humboldt, 1821) (Swarça et al., 2001), in different species of the genus Pimelodus (Souza et al., 2004; Garcia and Moreira-Filho, 2008), in Pseudoplatystoma corruscans (Spix and Agassiz, 1829) (Swarça et al., 2005), in Steindachneridion melanodermatum Garavello, 2005 and Steindachneridion scriptum (Miranda Ribeiro, 1918) (Swarça et al., 2008). In P. corruscans the 5S rDNA probe has been used in addition to the 18S rDNA showing that these regions are also present on different chromosomes in this species and located in a subterminal region (Swarça et al., 2005).
Martins and Galetti-Júnior (1999) described the location of the 45S and 5S rDNA regions on different chromosomes, which seems to represent a good condition, whereby the harmony of these sites of multiple copies would not be disrupted, which would be important to avoid undesirable translocations of 5S rDNA into the arrangement of 45S rDNA, or vice versa. Therefore, this condition allows an even greater conservatism of these regions, which is important for maintaining the cell.
Because 5S rDNA is found in the interstitial position of most fish of various orders, Martins and Galetti-Júnior (2001) suggest that this location is optimal in fish. Since the 5S rDNA region is located on the same chromosome in the populations of I. labrosus from the Capivara Reservoir (Carvalho and Dias, 2007) and from the Tibagi River (this study), but in terminal and interstitial positions, respectively, it is possible that there was a paracentric inversion in a subtelocentric pair bearing 5S ribosomal genes. Probably, due to the fact that in most fish species these genes are located in an interstitial position, the location of the terminal 5S genes in the population from Capivara represents a derived condition. According to Galetti-Júnior and Martins (2004), the interstitial chromosomal location does not seem to be coincidental, since this same pattern is observed in mammals and amphibians. This interstitial distribution may represent some advantage related to the organisation of these genes in the genome of vertebrates.
A pair of subtelocentric chromosomes in I. labrosus showed CMA3 positive sites on the long arm, probably indicating AgNOR, while with the fluorochrome DAPI these regions showed negative staining and were therefore rich in GC bases. Regions with similar CMA3 positive staining were also seen in the chromosomes of I. labrosus from the Capivara Reservoir (Carvalho and Dias, 2007).
Carvalho et al. (2004) described the supernumerary chromosomes of I. labrosus from the Tibagi River as totally heterochromatic. The results found in this study through the use of fluorochromes showed a particular heterochromatin in this supernumerary chromosome, with an abundance of GC. This was not seen in the population of I. labrosus from the Capivara Reservoir studied by Carvalho and Dias (2005a).
Few investigators have reported on the base composition of supernumerary chromosomes in fish; however, results similar to those obtained here were published by Vênere et al. (1999). These authors treated the chromosomes of Leporinus sp. with CMA3 and detected a small interstitial bright fluorescent band in the supernumerary chromosome of this species, which according to the authors may be related to a particular heterochromatin. Similarly, Abucarma and Martins-Santos (2001) reported an intense positive CMA3 signal on the metacentric B chromosome of Rhamdia branneri Haseman, 1911, in the telomeric heterochromatic region.
The findings of the present study suggest that, although there are some more conserved cytogenetic characteristics, such as the diploid number and NOR location, populations of I. labrosus may show their own characteristics. The karyotype formula, chromosomal location of 5S rDNA, and base composition of supernumerary chromosome of I. labrosus populations from the Tibagi River (current work) and from the Capivara Reservoir (Carvalho and Dias, 2005a, b, 2007) suggest that these populations are subject to different genetic changes that can be selected by the environment and may be of different species, thus requiring further taxonomic studies.
Acknowledgements - The authors are grateful to Fundação Araucária, CNPq (Conselho nacional de Desenvolvimento Científico e Tecnológico) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) for their financial support and to Dra. Maria Múlhmann Díaz (in memorian), who made available the Laboratório de Citogenética Molecular, CONEA, Buenos Aires.
Received April 24, 2009
Accepted October 5, 2009
Distributed August 31, 2010
- ABUCARMA, M. and MARTINS-SANTOS, IC., 2001. Karyotype and B chromosome of Rhamdia species (Pisces, Pimelodidae) endemic in the river Iguaçu basin. Cytologia, vol. 66, no. 3, p. 299-306.
- BERTOLLO, LAC., TAKAHASHI, CS. and MOREIRA FILHO, O., 1978. Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Brazilian Journal of Genetics, vol. 1, no. 2, p. 103-120.
- BORIN, LA. and MARTINS-SANTOS, IC., 2004. Study on karyotype and occurrence of B chromosomes in two endemic species of the genus Pimelodus (Siluriformes, Pimelodidae) from the River Iguaçu. Hereditas, vol. 140, p. 201-209.
- BURGESS, WE. 1989. An atlas of freshwater and marine catfishes: a preliminary survey of the Siluriformes. New Jersey: TFH Publications. 784 p.
- CARVALHO, RA., 2008. Ocorrência de cromossomos B em peixes neotropicais e análise citogenética em Iheringichthys labrosus e Megalonema platanum (Teleostei, Pimelodidae) Maringá: Universidade Estadual de Maringá. [Tese de Doutorado]
- CARVALHO, RA. and DIAS, AL., 2005a. Cytogenetic characterization of B chromosomes in two populations of Iheringichthys labrosus (Pisces, Pimelodidae) from the Capivara Reservoir (Paraná, Brazil). Caryologia, vol. 58, no. 3 p. 269-273.
- CARVALHO, RA. and DIAS, AL., 2005b. Karyotipic characterization of Iheringichthys labrosus (Pisces, Pimelodidae): C-, G- and restriction endonuclease banding. Genetics and Molecular Research, vol. 4, no. 4, p. 663-667.
- CARVALHO, RA. and DIAS, AL., 2007. Interindividual size heteromorphism of NOR and chromosomal location of 5S rRNA genes in Iheringichthys labrosus Brazilian Archives of Biology and Technology, vol. 50, no. 1, p. 141-146.
- CARVALHO, RA., GIULIANO-CAETANO, L. and DIAS, AL., 2004. Cytogenetic analysis of A- and B- chromosomes of Iheringichthys labrosus (Pisces, Pimelodidae) from the river Paraná, Brazil. Cytologia, vol. 69, no. 4, p. 381-385.
- CARVALHO, RA., MARTINS-SANTOS, IC. and DIAS, AL., 2008. B-chromosomes: an update about their occurrence in freshwater Neotropical fishes (Teleostei). Journal of Fish Biology, vol. 72, p. 1907-1932.
- CHRISTIAN, A., MCNIEL, E., ROBINSON, J., DRABEK, R., LARUE, S., WALDREN, C. and BEDFORD, J., 1998. A versatile image analysis approach for simultaneous chromosome identification and localization of FISH probes. Cytogenetics and Cell Genetics, vol. 82, no. 3-4, p. 172-179.
- DIAS, AL. and FORESTI, F., 1990. Algumas considerações sobre o cariótipo de Iheringichthys labrosus (Siluriformes, Pimelodidae) do Rio Mogi-Guaçu. In Abstract do III Simpósio de Citogenética Evolutiva Aplicada a Peixes Neotropicais Botucatu. p. 32.
- DIAS, AL. and FORESTI, F., 1993 Cytogenetic studies on fishes of the family Pimelodidae (Siluroidei). Brazilian Journal of Genetics, vol. 16, no. 3, p. 585-600.
- FENOCCHIO, AS., BERTOLLO, LAC., TAKAHASHI, CS., DIAS, AL. and SWARÇA, AC., 2003. Cytogenetic studies and correlated considerations on Rhamdiinae relationships (Pisces, Siluroidei, Pimelodidae). Cytologia, vol. 68, no. 4, p. 363-368.
- GALETTI JÚNIOR, PM. and MARTINS, C., 2004. Contribuição da hibridização in situ para o conhecimento dos cromossomos de peixes. In GUERRA, M. (Ed.). FISH: conceitos e aplicações na Citogenética. Ribeirão Preto: Sociedade Brasileira de Genética. p. 61-88.
- GARCIA, RMG., SACHETE, R. and MARTINS-SANTOS, IC., 1990. Aspectos citogenéticos de Iheringichthys labrosus (Pisces, Pimelodidae) do Rio Paraná, região de Porto Rico, PR. In Abstract do III Simpósio de Citogenética Evolutiva Aplicada a Peixes Neotropicais Botucatu. p. 33.
- GARCIA, C. and MOREIRA FILHO, O., 2008. Localization of ribosomal genes in three Pimelodus species (Siluriformes, Pimelodidae) of the São Francisco River: 5S genes as species markers and conservation of the 18S rDNA sites. Genetics and Molecular Biology, vol. 31, no. (suppl), p. 261-264.
- HOWELL, WM. and BLACK, DA., 1980. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: a one-step method. Experientia, vol. 36, no. 8, p. 1014-1015.
- JESUS, CM., GALETTI JÚNIOR, PM., VALENTINI, S. and MOREIRA FILHO, O., 2003. Molecular characterization and chromosomal localization of two families of satellite DNA in Prochilodus lineatus (Pisces, Prochilodontidae), a species with B chromosomes. Genetica, vol. 118, no. 1, p. 25-32.
- MAISTRO, EL., OLIVEIRA, C. and FORESTI, F., 2000. Cytogenetic analysis of A- and B-chromosomes of Prochilodus lineatus (Teleostei, Prochilodontidae) using different restriction enzyme banding and staining methods. Genetica, vol. 108, no. 2, p. 119-125.
- MAISTRO, EL., OLIVEIRA, C. and FORESTI, F., 2002. Cytogenetic analysis of A- and B-chromosomes of Rhamdia hilarii (Teleostei, Pimelodidae): C-Banding, silver nitrate and CMA3 staining and restriction endonucleases banding. Cytologia, vol. 67, no. 1, p. 25-31.
- MARTINS, C. and GALETTI JÚNIOR, PM., 1999. Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Research, vol. 7, no. 5, p. 363-367.
- MARTINS, C. and GALETTI JÚNIOR, PM., 2001. Two 5S rDNA arrays in Neotropical fish species: Is it a rule for fishes? Genetica, vol. 111, no. 1-3, p. 439-446.
- RIBEIRO, LB., MATOSO, DA., ALMEIDA, MC., VICARI, MR., MORAES NETO, A., SVIDNICKI, MC. and ARTONI, RF., 20068. Karyotypic variability in Iheringichthys labrosus (Teleostei, Pimelodidae) from the Tibagi River basin (Paraná State, Brazil). Genetics and Molecular Research, vol. 7, no. 3, p. 718-724.
- SCHWEIZER, D., 1980. Simultaneous fluorescent staining of R bands and specific heterochromatic regions (DA/DAPI) in human chromosomes. Cytogenetics and Cell Genetics, vol. 27, no. 2-3, p. 190-193.
- SOUZA, L., GIULIANO-CAETANO, L. and DIAS, AL., 2004. Banding chromosome pattern of two species of Pimelodus (Siluriformes, Pimelodidae) from the Paraná river basin of Brazil. Folia Biologica, vol. 52, no. 3-4, p. 165-169.
- SWARÇA, AC., CESTARI, MM., GIULIANO-CAETANO, L. and DIAS, AL., 2001. Cytogenetic characterization of the large South American siluriform fish species Zungaro zungaro (Pisces, Pimelodidae). Chromosome Science, vol. 5, no. 1, p. 51-55.
- SWARÇA, AC., FENOCCHIO, AS., CESTARI, MM. and DIAS, AL., 2003. Analysis of heterochromatin by combination of C-banding and CMA3 and DAPI staining in two fish species (Pimelodidae, Siluriformes). Genetica, vol. 119, no. 1, p. 87-92.
- SWARÇA, AC., FENOCCHIO, AS., CESTARI, MM. and DIAS, AL., 2005. Karyotype divergence among populations of giant catfish Pseudoplatystoma corruscans (Teleostei: Pimelodidae) indicates higher species diversity. Ichthyological Exploration of Freshwaters, vol. 16, no. 4, p. 325-330.
- SWARÇA, AC., FENOCCHIO, AS., CESTARI, MM. and DIAS, AL., 2008. Analyses of the structure of NORs in two species of South American Sorubiminae fishes (Siluriformes) by means of several cytogenetic techniques. Folia Biologica, vol. 56, no. 1-2, p. 31-35.
- SWARÇA, AC., GIULIANO-CAETANO, L. and DIAS, AL., 1999. Cytogenetic characterization through chromosomic banding of Pinirampus pirinampu (Pisces, Pimelodidae) from the Tibagi river basin PR/Brazil. Caryologia, vol. 52, no. 1-2, p. 31-35.
- TRECO, FR., MALABARBA, LR., GIULIANO-CAETANO, L. and DIAS, AL., 2008. Cytogenetic study of two species of the family Pimelodidae (Siluriformes) collected in lago Guaíba, Rio Grande do Sul, Brazil. Neotropical Ichthyology, vol. 6, no. 1, p. 87-92.
- VÊNERE, PC., MIYAZAWA, CS. and GALETTI JÚNIOR, PM., 1999. New cases of supernumerary chromosomes in characiform fishes. Genetics and Molecular Biology, vol. 22, no. 3, p. 345-349.
- VISSOTTO, PC., FORESTI, F. and OLIVEIRA, C., 1999. Supernumerary chromosomes in two species of the family Pimelodidae (Teleostei, Siluriformes). Chromosome Science, vol. 3, no. 1, p. 9-13.
Publication Dates
-
Publication in this collection
16 Aug 2010 -
Date of issue
Aug 2010
History
-
Received
24 Apr 2009 -
Accepted
05 Oct 2009