Abstract
Alpha amylase, catalyzing the hydrolysis of starch is a ubiquitous enzyme with tremendous industrial applications. A 1698 bp gene coding for 565 amino acid amylase was PCR amplified from Geobacillus thermodenitrificans DSM-465, cloned in pET21a (+) plasmid, expressed in BL21 (DE3) strain of E. coli and characterized. The recombinant enzyme exhibited molecular weight of 63 kDa, optimum pH 8, optimum temperature 70°C, and KM value of 157.7µM. On pilot scale, the purified enzyme efficiently removed up to 95% starch from the cotton fabric indicating its desizing ability at high temperature. 3D model of enzyme built by Raptor-X and validated by Ramachandran plot appeared as a monomer having 31% α-helices, 15% β-sheets, and 52% loops. Docking studies have shown the best binding affinity of enzyme with amylopectin (∆G -10.59). According to our results, Asp 232, Glu274, Arg448, Glu385, Asp34, Asn276, and Arg175 constitute the potential active site of enzyme.
Keywords:
amylase; Geobacillus; cloning; properties; in silico
Resumo
A alfa-amilase, que catalisa a hidrólise do amido, é uma enzima ubíqua com imensas aplicações industriais. Um gene de 1698 pb que codifica a amilase de 565 aminoácidos foi amplificado por PCR, a partir de Geobacillus thermodenitrificans DSM-465, clonado no plasmídeo pET21a (+), expresso na cepa BL21 (DE3) de E. coli e caracterizado. A enzima recombinante exibiu peso molecular de 63 kDa, pH ótimo igual a 8, temperatura ótima de 70° C e valor KM de 157,7 µM. Em escala piloto, a enzima purificada removeu com eficiência até 95% de amido do tecido de algodão, indicando sua capacidade de desengomagem em alta temperatura. O modelo 3D da enzima construída por Raptor-X e validada por Ramachandran plot apareceu como um monômero com 31% de hélices alfa, 15% de folhas beta e 52% de loops. Os estudos de docking mostraram melhor afinidade de ligação da enzima com amilopectina (∆G: - 10,59). De acordo com nossos resultados, Asp 232, Glu274, Arg448, Glu385, Asp34, Asn276 e Arg175 constituem o sítio ativo potencial da enzima.
Palavras-chave:
amilase; Geobacillus; clonagem, propriedades; in sílico
1. Introduction
Alpha-amylase is a hydrolytic enzyme of primary metabolism that catalyses the breakdown of α-1,4-glycosidic linkage of glucose in starch, liberating glucose and/or maltose. It is a metaloenzyme that can be either exohydrolase (Gupta et al., 2003GUPTA, R., GIGRAS, P., MOHAPATRA, H., GOSWAMI, V.K. and CHAUHAN, B., 2003. Microbial α-amylases: a biotechnological perspective. Process Biochemistry, vol. 38, no. 11, pp. 1599-1616. http://dx.doi.org/10.1016/S0032-9592(03)00053-0.
http://dx.doi.org/10.1016/S0032-9592(03)...
; Sundarram and Murthy, 2014SUNDARRAM, A. and MURTHY, T.P.K., 2014. α-amylase production and applications: a review. Journal of Applied & Environmental Microbiology, vol. 2, no. 4, pp. 166-175.) or endohydrolase (Yang et al., 2017YANG, G., YAO, H., MOZZICAFREDDO, M., BALLARINI, P., PUCCIARELLI, S. and MICELI, C., 2017. Rational engineering of a cold-adapted α-amylase from the Antarctic ciliate Euplotes focardii for simultaneous improvement of thermostability and catalytic activity. Applied and Environmental Microbiology, vol. 83, no. 13, pp. e00449-17. http://dx.doi.org/10.1128/AEM.00449-17. PMid:28455329.
http://dx.doi.org/10.1128/AEM.00449-17...
). Majority of α-amylases have two aspartate and one glutamate amino acids that play an essential role in catalysis (Sarian et al., 2017SARIAN, F.D., JANEČEK, Š., PIJNING, T., NURACHMAN, Z., RADJASA, O.K., DIJKHUIZEN, L., NATALIA, D. and VAN DER MAAREL, M.J., 2017. A new group of glycoside hydrolase family 13 α-amylases with an aberrant catalytic triad. Scientific Reports, vol. 71, pp. 1-10.). The α-amylases can have eukaryotic archaeal, or bacterial origin, the latter having highest diversity and being at the higher evolutionary distance from the eukaryotic enzymes. Physiologically, the α-amylases play an important role in the digestion of starch in the human body (Mehta and Satyanarayana, 2016MEHTA, D. and SATYANARAYANA, T., 2016. Bacterial and archaeal α-amylases: diversity and amelioration of the desirable characteristics for industrial applications. Frontiers in Microbiology, vol. 7, pp. 1129. http://dx.doi.org/10.3389/fmicb.2016.01129. PMid:27516755.
http://dx.doi.org/10.3389/fmicb.2016.011...
; Yan and Wu, 2017YAN, S. and WU, G., 2017. Bottleneck in secretion of α-amylase in Bacillus subtilis. Microbial Cell Factories, vol. 16, no. 1, pp. 124. http://dx.doi.org/10.1186/s12934-017-0738-1. PMid:28724440.
http://dx.doi.org/10.1186/s12934-017-073...
; Ahmed et al., 2021AHMED, S., ZIA, A., MEHMOOD, S.A., PANHWAR, W.A., KHAN, W., SHAH, M., ULLAH, I., 2021. Change in malate dehydrogenase and alpha amylase activities in Rubus fruticosus and Valeriana jatamansi treated granary weevil, Sitophilus granarius. Brazilian Journal of Biology = Revista Brasileira de Biologia, vol. 81, no. 2, pp. 387-391. http://dx.doi.org/10.1590/1519-6984.226952.
https://doi.org/10.1590/1519-6984.226952...
). The α-amylases are important products obtained from the living organisms that have a wide range applications in clinical, pharmaceutical and other industries. These days, the chemical hydrolysis of starch on the industrial scale has been replaced by amylase catalyzed processing. Amylases have great value in the textile, fermentation, food, paper and other industries (Pandey et al., 2000PANDEY, A., NIGAM, P., SOCCOL, C.R., SOCCOL, V.T., SINGH, D. and MOHAN, R., 2000. Advances in microbial amylases. Biotechnology and Applied Biochemistry, vol. 31, no. 2, pp. 135-152. http://dx.doi.org/10.1042/BA19990073. PMid:10744959.
http://dx.doi.org/10.1042/BA19990073...
). This is the most frequently used industrial enzyme which counts about 25% of total enzyme market worldwide (Sidhu et al., 1997SIDHU, G., SHARMA, P., CHAKRABARTI, T. and GUPTA, J., 1997. Strain improvement for the production of a thermostable α-amylase. Enzyme and Microbial Technology, vol. 21, no. 7, pp. 525-530. http://dx.doi.org/10.1016/S0141-0229(97)00055-0.
http://dx.doi.org/10.1016/S0141-0229(97)...
; Rao et al., 1998RAO, M.B., TANKSALE, A.M., GHATGE, M.S. and DESHPANDE, V.V., 1998. Molecular and biotechnological aspects of microbial proteases. Microbiology and Molecular Biology Reviews, vol. 62, no. 3, pp. 597-635. http://dx.doi.org/10.1128/MMBR.62.3.597-635.1998. PMid:9729602.
http://dx.doi.org/10.1128/MMBR.62.3.597-...
). Amylase is an essential ingredient in the current vegetable, fruit and juice industries to aid juicing, peeling, to make the clear extract and improve the quality (Kumar, 2015KUMAR, S., 2015. Role of enzymes in fruit juice processing and its quality enhancement. Health, vol. 6, no. 6, pp. 114-124.; Garg et al., 2016GARG, G., SINGH, A., KAUR, A., SINGH, R., KAUR, J., MAHAJAN, R., 2016. Microbial pectinases: an ecofriendly tool of nature for industries. 3 Biotech, vol. 6, no. 1, pp. 47.). In particular, the cloudy and thick juices are clarified by the application of amylases along with pectinases and cellulases (Allan et al., 1997ALLAN, S., TORBENVEDEL, B. and HENRICK, B., 1997. Recombinant alpha amylase mutants and their use in textile desizing starch liquification and washing. PTC International Patent Application, vol. 12, no. 5, pp. 205-210.; Sivaramakrishnan et al., 2006SIVARAMAKRISHNAN, S., GANGADHARAN, D., NAMPOOTHIRI, K.M., SOCCOL, C.R. and PANDEY, A., 2006. Alpha - Amylases from microbial sources–an overview on recent developments. Food Technology and Biotechnology, vol. 44, no. 2, pp. 173-184.). Starch paste is applied in warping in textile industries during weaving, to give strength to the thread and avoids the loss of strings by friction. The starch layer is removed to make the cloth smooth by the application of amylase. Alpha-amylase is applied in textile industries for desizing and removing starch before dying (Asgher et al., 2007ASGHER, M., ASAD, M.J., RAHMAN, S.U. and LEGGE, R.L., 2007. A thermostable α-amylase from a moderately thermophilic Bacillus subtilis strain for starch processing. Journal of Food Engineering, vol. 79, no. 3, pp. 950-955. http://dx.doi.org/10.1016/j.jfoodeng.2005.12.053.
http://dx.doi.org/10.1016/j.jfoodeng.200...
; Zafar et al., 2019ZAFAR, A., AFTAB, M.N., IQBAL, I., DIN, Z. and SALEEM, M.A., 2019. Pilot-scale production of a highly thermostable α-amylase enzyme from Thermotoga petrophila cloned into E. coli and its application as a desizer in textile industry. RSC Advances, vol. 9, no. 2, pp. 984-992. http://dx.doi.org/10.1039/C8RA06554C.
http://dx.doi.org/10.1039/C8RA06554C...
). The bacterial enzymes have a wide optimal temperature range for their activity starting from less than ∼25°C to around 100°C. Similarly, the pH for optimum activity of α-amylases of bacterial origin can be from 1 to 11.5 (Yan and Wu, 2017YAN, S. and WU, G., 2017. Bottleneck in secretion of α-amylase in Bacillus subtilis. Microbial Cell Factories, vol. 16, no. 1, pp. 124. http://dx.doi.org/10.1186/s12934-017-0738-1. PMid:28724440.
http://dx.doi.org/10.1186/s12934-017-073...
). These properties make amylases from moderately thermophilic bacteria more suitable to be used at industrial temperatures (Ghasemi et al., 2015GHASEMI, A., GHAFOURIAN, S., VAFAEI, S., MOHEBI, R., FARZI, M., TAHERIKALANI, M. and SADEGHIFARD, N., 2015. Cloning, Expression, and Purification of Hyperthermophile α-Amylase from Pyrococcus woesei. Osong Public Health and Research Perspectives, vol. 6, no. 6, pp. 336-340. http://dx.doi.org/10.1016/j.phrp.2015.10.003. PMid:26835242.
http://dx.doi.org/10.1016/j.phrp.2015.10...
; Saini et al., 2017SAINI, R., SAINI, H.S. and DAHIYA, A., 2017. Amylases: characteristics and industrial applications. Journal of Pharmacognosy and Phytochemistry, vol. 6, no. 4, pp. 1865-1871.). Recombinant DNA technology has made it possible to fulfil the ever increasing demands of amylase in the industries (Zeigler, 2014ZEIGLER, D. R., 2014. The Geobacillus paradox: why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology, vol. 160, pp. 1-11. https://doi.org/10.1099/mic.0.071696-0.
https://doi.org/10.1099/mic.0.071696-0...
). Recombinant microbial sources more satisfy the industrial demands since last three decades. Geobacillus is a genus including aerobic and facultatively anaerobic, rod-shaped, spore-forming microbes. The members of this group can grow at a wide range of temperatures from 35oC to 80oC. However, of the species grow at the temperature range between 45°C and 70°C. These features make Geobacilli the attractive source of enzymes and proteins produced in the biotechnology industry (Wang et al., 2019WANG, G., LUO, M., LIN, J., LIN, Y., YAN, R., STREIT, W.R. and YE, X., 2019. A new extremely halophilic, calcium-independent and surfactant-resistant alpha-amylase from Alkalibacterium sp. SL3. Journal of Microbiology and Biotechnology, vol. 29, no. 5, pp. 765-775. http://dx.doi.org/10.4014/jmb.1901.01038. PMid:30982319.
http://dx.doi.org/10.4014/jmb.1901.01038...
). The present research describes the biochemical and in silico studies of recombinant alpha-amylase from Geobacillus thermodenitrificans DSM 465.
2. Materials and Methods
2.1. Materials
All the chemicals and kits for PCR, DNA restriction, cloning, plasmid isolation, gel DNA elution, plasmid restriction, purification and characterization of enzyme were obtained from Thermo and Sigma-Aldrich. The genomic DNA of a bacterial strain to be used as the source of genes were obtained from DSMZ Germany.
2.2. PCR amplification and cloning
The gene comprising of about 1700 bp was PCR amplified using the primers sequence 5’-gcatatgaggaaacaaactatattgccgca-3’and 5’-aatgaattctcatcgatccgccttgac-3’ with NdeI and EcoRI restriction sites. The gene was ligated into the pTZ57R/T plasmid and bacterial cells were transformed using the procedure provided by Thermo Scientific. InsTAclone PCR Cloning Kit. #K1214). Successful transformants were initially selected on the basis of blue-white colonies on LB agar medium containing 100µg/ml ampicillin, 0.5mM β-D1-thiogalactopyranoside (IPTG), and 50mg/ml of 5-Bromo-4-chloro-3-indolyl β-D-galactopyranoside (x-gal). The plasmids were confirmed by restriction analysis. The amylase gene restricted out from pTZ-amy+ as two fragments as there is a restriction site for NdeI at about 880bp downstream to the start codon, both of the fragments were purified from the agarose gel, ligated and subcloned into pET21a (+) plasmid restricted with the same pair of enzymes to prepare recombinant plasmid pET21-amy+.
2.3. Expression of amylase gene in E. coli
E. coli cells BL21 (DE3) cells were transformed by ligation mixture containing the recombinant plasmid pET21-amy+. An individual bacterial colony was grown in the LB broth medium supplemented with ampicillin (100 μg/mL) in an incubating shaker adjusted at 200 rpm and 37°C. The enzyme was produced in the medium containing 0.5mM IPTG incubated for 3.5 hours. Bacterial cells transformed with a plasmid pET21-amy- was grown and analyzed as the negative control.
2.4. Purification and kinetic properties of amylase
The bacterial culture was centrifuged at 7000 x g for 5 min, precipitate was resuspended in the buffer solution A (20mM Tris-HCl pH 8.0). The cells were sonicated, and clear supernatant was obtained by centrifugation at 12,000 × g. The activity of enzyme was measured in the clear solution and further processed for enzyme purification by selective precipitation with chilled ethanol followed by dialysis and DEAE-Sephadex chromatography. The enzyme sample dialyzed in the same buffer was loaded onto the colum at a flow rate of 2ml per minute. The protein contaminations were washed with buffer A and target protein was eluted by a linear NaCl gradient 0 to 0.5M. Fractions containing 3 mL of enzyme sample were collected manually and further analysed.
The enzyme activity was measured by a method based on dinitrosalicylic acid (DNS) (Wang et al., 2019WANG, G., LUO, M., LIN, J., LIN, Y., YAN, R., STREIT, W.R. and YE, X., 2019. A new extremely halophilic, calcium-independent and surfactant-resistant alpha-amylase from Alkalibacterium sp. SL3. Journal of Microbiology and Biotechnology, vol. 29, no. 5, pp. 765-775. http://dx.doi.org/10.4014/jmb.1901.01038. PMid:30982319.
http://dx.doi.org/10.4014/jmb.1901.01038...
; Zafar et al., 2019ZAFAR, A., AFTAB, M.N., IQBAL, I., DIN, Z. and SALEEM, M.A., 2019. Pilot-scale production of a highly thermostable α-amylase enzyme from Thermotoga petrophila cloned into E. coli and its application as a desizer in textile industry. RSC Advances, vol. 9, no. 2, pp. 984-992. http://dx.doi.org/10.1039/C8RA06554C.
http://dx.doi.org/10.1039/C8RA06554C...
). Standard curve for known maltose concentrations was prepared. The enzyme dilution (50 to 100 µL) was mixed with 2 mL of starch solution prepared in 50mM buffer A. adjusted at 70°C and incubated for 5 min. The reaction mixture was added with 1 mL of DNS reagent and incubated for 5 min in the boiling water in a water bath. The amount of maltose was measured by absorbance at 540 nm. The amount of enzyme that can liberate one micromole of reducing sugar from starch under our assay conditions was considered as one unit. The enzyme stability and activity was determined at different pH levels (4.5 to 10) and temperature ranging from 40°C to 90°C. Total protein content of final solution was determined by Bradford method (Bradford, 1976BRADFORD, M.M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, vol. 72, pp. 248-254. http://dx.doi.org/10.1016/0003-2697(76)90527-3. PMid:942051.
http://dx.doi.org/10.1016/0003-2697(76)9...
). The enzyme activity was measured under linear increase in the starch concentration, Lineweaver-burk plot procedure was used to determined KM and Vmax values.
2.5. Textile desizing ability of enzyme
The starch removing ability of enzyme was determined by using a piece of cotton grey cloth coated with starch paste. The cloth was subjected to thermal aging at 80°C for 10 days solidification. The cloth, coated with starch was subjected to boiling water for one to two minutes to activate the starch layer. Three cotton cloths (5x8 cm) coated with starch were used in the study. Pre and post-treatment weight of starch coated cloth was determined. The material was immersed for 1 hour in 100 mL aqueous buffer solution (100 mM Tris-HCl, pH 8) adjusted at 70°C containing 200 U of α-amylase. The fabric strip was dried in the oven post enzyme treatment and its weight was determined. The weight of starch removed was calculated by subtracting the final weight from initial weight of the cloth. The amount of hydrolysed starch was assessed after enzymic treatment of fabric cloth by determination of maltose using DNS (3, 5-dinitrosalicylic acid) reagent method (Saha et al., 2018SAHA, P., KHAN, M.F. and PATRA, S., 2018. Truncated α-amylase: an improved candidate for textile processing. Preparative Biochemistry & Biotechnology, vol. 48, no. 7, pp. 635-645. http://dx.doi.org/10.1080/10826068.2018.1479863. PMid:29902115.
http://dx.doi.org/10.1080/10826068.2018....
). The changes in weight of pre and post treated cloth was also used for the determination of starch by using following formula 1:
Where, W1 is weight (mg) of fabric before application of starch, W2 of weight of fabric with starch paste applied, W3 is weight of cloth after enzyme treatment.
2.6. In silico 3-D structure determination of amylase
The 3-dimensional structure of the protein was generated using I-TASSER (Yang and Zhang, 2015YANG, J. and ZHANG, Y., 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Research, vol. 43, no. W1, pp. W174-W181. http://dx.doi.org/10.1093/nar/gkv342. PMid:25883148.
http://dx.doi.org/10.1093/nar/gkv342...
), Swiss-Model (Waterhouse et al., 2018WATERHOUSE, A., BERTONI, M., BIENERT, S., STUDER, G., TAURIELLO, G., GUMIENNY, R., HEER, F.T., DE BEER, T.A.P., REMPFER, C., BORDOLI, L., LEPORE, R. and SCHWEDE, T., 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research, vol. 46, no. W1, pp. W296-W303. http://dx.doi.org/10.1093/nar/gky427. PMid:29788355.
http://dx.doi.org/10.1093/nar/gky427...
), and Raptor-X (Källberg et al., 2012KÄLLBERG, M., WANG, H., WANG, S., PENG, J., WANG, Z., LU, H. and XU, J., 2012. Template-based protein structure modeling using the RaptorX web server. Nature Protocols, vol. 7, no. 8, pp. 1511-1522. http://dx.doi.org/10.1038/nprot.2012.085. PMid:22814390.
http://dx.doi.org/10.1038/nprot.2012.085...
) servers, these programs were used to generate the best quality score model, due to the absence of a significant match (27%) with structure available in the PDB database. The quality score of the model built was validated by server quality parameters and Ramachandran plot (Furnham et al., 2006FURNHAM, N., DORÉ, A.S., CHIRGADZE, D.Y., DE BAKKER, P.I., DEPRISTO, M.A. and BLUNDELL, T.L., 2006. Knowledge-based real-space explorations for low-resolution structure determination. Structure (London, England : 1993), vol. 14, no. 8, pp. 1313-1320. http://dx.doi.org/10.1016/j.str.2006.06.014. PMid:16905105.
http://dx.doi.org/10.1016/j.str.2006.06....
). The model generated by Raptor-X was found to have best quality, which has been selected for docking.
2.7. Molecular docking analysis
Molecular Operating Environment (MOE) software was applied for molecular docking studies (Sapundzhi, F.I., Dzimbova, 2018SAPUNDZHI, F.I. and DZIMBOVA, T.A., 2018. Computer modelling of the CB1 receptor by molecular operating environment. Bulgarian Chemical Communications, vol. 50. Special Issue B, pp. 15-19.). The selected ligands were analysed, energy was minimized and all atoms were partially charged, protein 3D structure automatically quickly prepared by MOE. The active site was determined and isolated by MOE site finder. The docking was based on minimum energy calculation, best fit poses in the cavity, and distance of hydrogen bonds between ligands and receptor. The best pose was selected for each ligand i.e. amylose, amylopectin, starch, glycogen and maltose etc. Chimera software (Pettersen et al., 2004PETTERSEN, E.F., GODDARD, T.D., HUANG, C.C., COUCH, G.S., GREENBLATT, D.M., MENG, E.C. and FERRIN, T.E., 2004. UCSF Chimera: a visualization system for exploratory research and analysis. Journal of Computational Chemistry, vol. 25, no. 13, pp. 1605-1612. http://dx.doi.org/10.1002/jcc.20084. PMid:15264254.
http://dx.doi.org/10.1002/jcc.20084...
) was used for visualization and calculations of ligands and protein interaction and estimation of H-bonds distances.
3. Results
3.1. Production, purification and kinetics of amylase
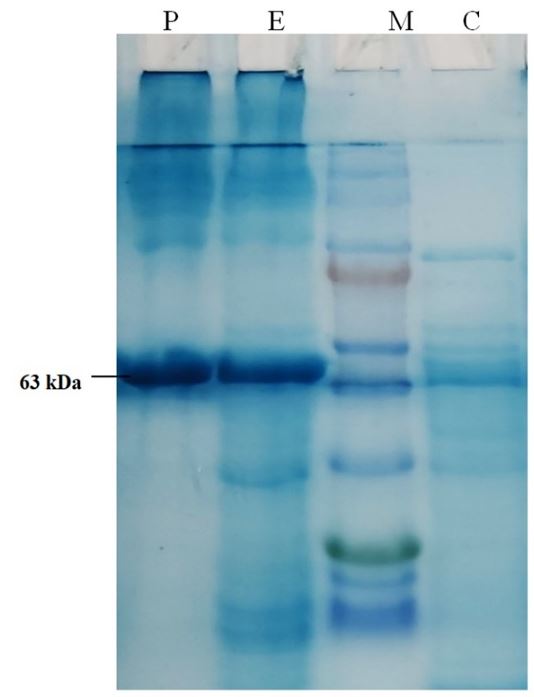
The recombinant alpha amylase was successfully produced in E. coli cells under T7 promoter system. It was purified and gave a protein band at about 63kDa on SDS-PAGE (Figure 1). The specific activity of purified enzyme was 153.6 U/mg, it was purified to 12.5 folds with 54.13% final recovery (Table 1). The optimum activity of amylase was measured at 70°C and pH 8. The KM value of enzyme was found 157.7µM, Vmax 333.3 micromoles/min, and Vmax/KM ratio was 2.11 (Figure 2).
SDS-PAGE photograph indicating the expression and purification of alpha amylase. Lane C- control experiment (without gene), Lane-M. Protein marker (ThermoFisher Scientific PageRulerTM Prestained Protein Ladder, 10 kDa to 180 kDa), Lane E, experimental with expression of gene, Lane P, partially purified enzyme alpha amylase. The molecular weight of the purified enzyme was found as 63 kDa on SDS-PAGE.
Enzyme activity, total protein content, specific activity, percentage yield and fold purification of recombinant amylase from Geobacillus thermodenitrificans DSM-465. The amount of enzyme that can liberate one micromole of maltose from starch under our assay conditions is known as one unit of enzyme.
Enzyme kinetics. (A) The effect of temperature on the enzyme stability; (B) determination of KM and Vmax values of purified alpha-amylase using Lineweaver-Burk plot; (C) The effect of reaction mixture temperature on the enzyme activity indicating an optimum temperature of 70°C; (D) The effect of pH on the enzyme activity, showing maximum activity at pH 8.
3.2. Textile fabric desizing by amylase
The textile desizing ability of recombinant enzyme was determined by using cotton fabric coated with starch paste. According to the weight estimation method, up to 95% starch was hydrolysed by the enzyme. There was no further increase in the liberated maltose concentration after 45 minutes of enzyme treatment indicating the maximum hydrolysis of starch.
3.3. 3D modelling of protein structure
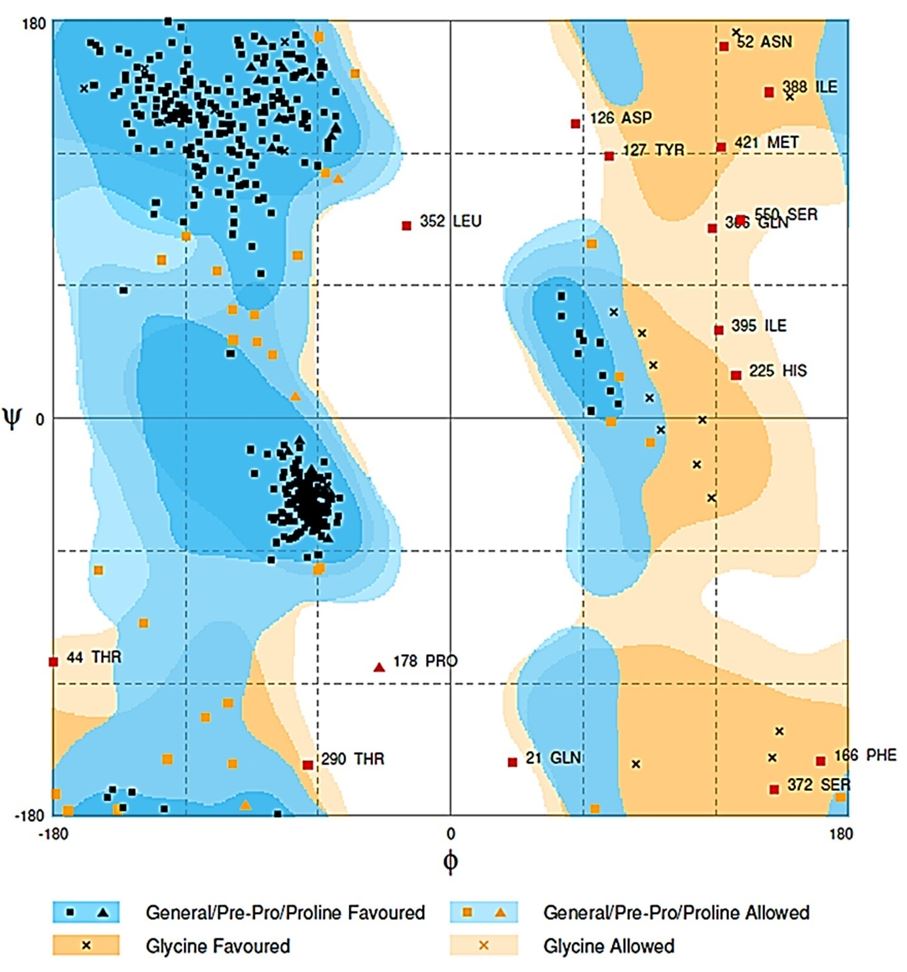
The best model was generated by Raptor-X server, the input was predicted as one domain, that match the best template 3ueqA, p-value 2.33e-14, all alpha-amylase 565 (100%) residues were modelled, 6 (1%) positions were predicted as disordered. Secondary structure comprised 31% of α-helix, 15% β-sheet, and 52% loop. The solvent access distribution indicated, 30% E residue, 27%M, and 41% B (Figure 3). I-TASSER model displayed about 98% residues in the expected region, 71.2% residues in allowed region, and 18.5% residues in outlier region. Swiss-Model model score confirmed 90.2%, 6.6%, and 3.2% residues in the expected regions in a model. Raptor-X model scored 91.5% residues in the expected region, 5.7% residues in allowed region and only 2.8% in the outer range. 3D structure of the protein was validated by Ramachandran plot (Figure 4). The conservation of amino acid residues in the protein structure was determined by ConSurf server (Celniker et al., 2013CELNIKER, G., NIMROD, G., ASHKENAZY, H., GLASER, F., MARTZ, E., MAYROSE, I., PUPKO, T. and BEN‐TAL, N., 2013. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Israel Journal of Chemistry, vol. 53, no. 3-4, pp. 199-206. http://dx.doi.org/10.1002/ijch.201200096.
http://dx.doi.org/10.1002/ijch.201200096...
).
3-dimentionsl protein structure of alpha-amylase built by Raptor-X software. α-helices are indicated by red, β-sheet by yellow and random coils by white. The protein structure consists of a single monomer.
Ramachandran plot for validation of alpha-amylase enzyme 3D structure generated by Raptor-X server.
3.4. Molecular docking analysis
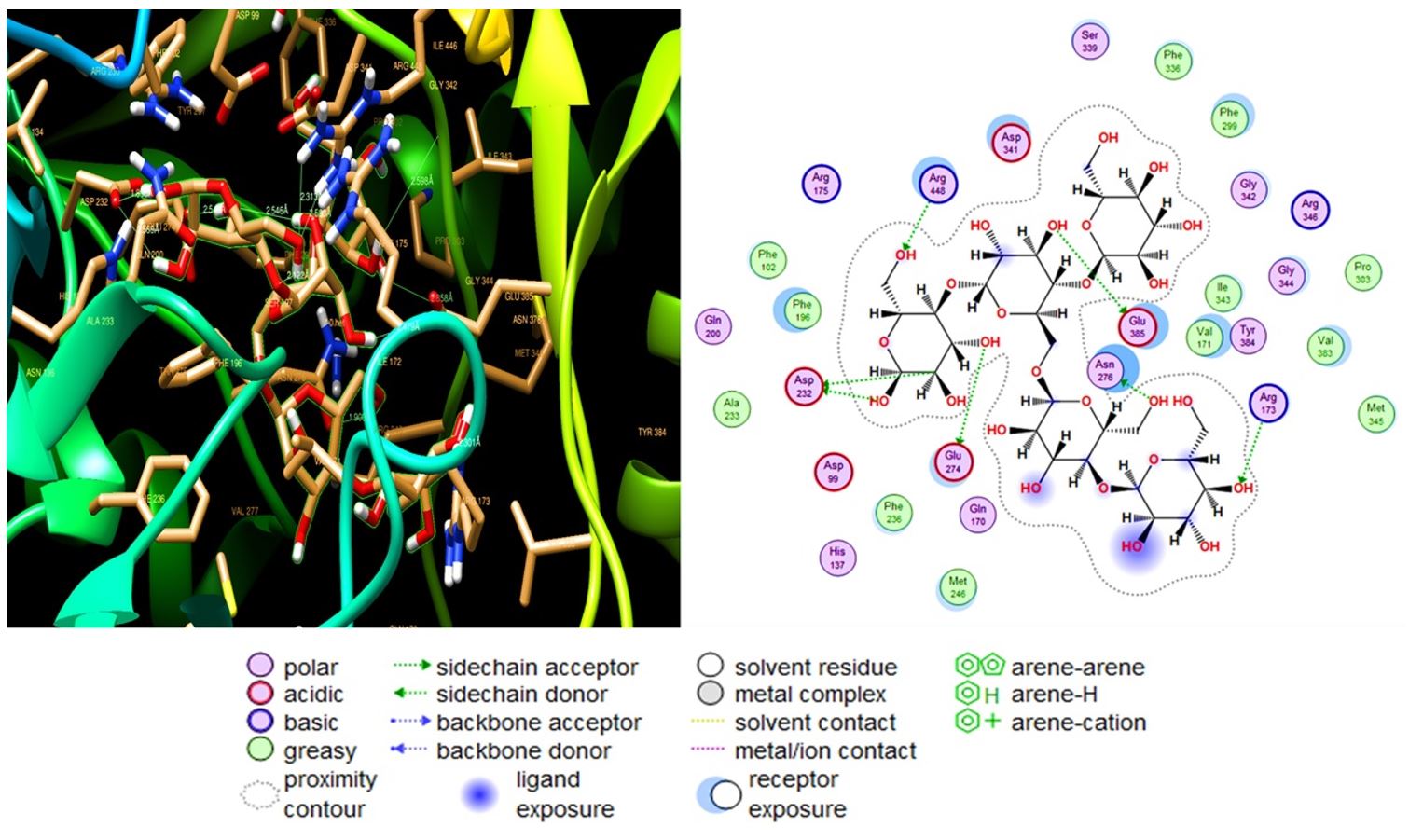
Five ligands were analysed for their binding affinities with 3D model of amylase. All of these molecules formed weak to strong hydrogen bonds (distance was less than 2.86Å) with Asp 232, Arg448, Glu385, Asn276 and, Arg173 residues. With the highest RMSD value (2.44). A pose with RMSD less than 2 Å are considered as the best suited for a structural model for dockings studies, the values between 2 to 3 Å are also considered as reliable models (Wang et al., 2016WANG S, LI W, LIU S, XU J., 2016. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Research, vol. 44, no. W1, pp. W430-W435.). Amylopectin interacted most strongly with protein with binding energy equal to ∆G -10.59 and formation of six side-chain H-bonds (distance 1.7 to 2.3 Å), as interacting amino acids (Figure 5). Glycogen interacted with protein scored ∆G -10.41, formed 9 side-chain H-bonds (distance 1.8 to 2.88 Å) (Supplementary Material Supplementary Material Supplementary material accompanies this paper Figure S1. The interaction of glycogen molecule (PubChem ID - 439177) and alpha-amylase enzyme active site. Glycogen was the second most strongly binding molecule with amylase structure. Figure S2. Molecular interaction of starch molecule (PubChem ID - 24836924) and alpha-amylase enzyme active site amino acids as determined by molecular docking analysis. Figure S3. Docking based interaction of amylose molecule (PubChem ID - 53477771) and alpha-amylase enzyme active site amino acids. Figure S4. Molecular docking results showing the interaction of maltose molecule (PubChem ID - 439341) and alpha-amylase enzyme active site amino acids. This material is available as part of the online article from http://www.scielo.br/BJB ). Starch strongly interacted with protein with binding energy equal to -10.04 and formation of six side-chain H-bonds (distance 1.7- 2.3 Å, (Supplementary material Supplementary Material Supplementary material accompanies this paper Figure S1. The interaction of glycogen molecule (PubChem ID - 439177) and alpha-amylase enzyme active site. Glycogen was the second most strongly binding molecule with amylase structure. Figure S2. Molecular interaction of starch molecule (PubChem ID - 24836924) and alpha-amylase enzyme active site amino acids as determined by molecular docking analysis. Figure S3. Docking based interaction of amylose molecule (PubChem ID - 53477771) and alpha-amylase enzyme active site amino acids. Figure S4. Molecular docking results showing the interaction of maltose molecule (PubChem ID - 439341) and alpha-amylase enzyme active site amino acids. This material is available as part of the online article from http://www.scielo.br/BJB ). Amylose scored ∆G -7.49 binding energy and formed ten side-chains H-bonds (distance 1.8-2.8 Å) (Supplementary material Supplementary Material Supplementary material accompanies this paper Figure S1. The interaction of glycogen molecule (PubChem ID - 439177) and alpha-amylase enzyme active site. Glycogen was the second most strongly binding molecule with amylase structure. Figure S2. Molecular interaction of starch molecule (PubChem ID - 24836924) and alpha-amylase enzyme active site amino acids as determined by molecular docking analysis. Figure S3. Docking based interaction of amylose molecule (PubChem ID - 53477771) and alpha-amylase enzyme active site amino acids. Figure S4. Molecular docking results showing the interaction of maltose molecule (PubChem ID - 439341) and alpha-amylase enzyme active site amino acids. This material is available as part of the online article from http://www.scielo.br/BJB ). Maltose scored the lowest binding energy (∆G -2.8) and showed five side-chain interacted H-bonds (distance 1.78 to 2.5 Å) (Supplementary material Supplementary Material Supplementary material accompanies this paper Figure S1. The interaction of glycogen molecule (PubChem ID - 439177) and alpha-amylase enzyme active site. Glycogen was the second most strongly binding molecule with amylase structure. Figure S2. Molecular interaction of starch molecule (PubChem ID - 24836924) and alpha-amylase enzyme active site amino acids as determined by molecular docking analysis. Figure S3. Docking based interaction of amylose molecule (PubChem ID - 53477771) and alpha-amylase enzyme active site amino acids. Figure S4. Molecular docking results showing the interaction of maltose molecule (PubChem ID - 439341) and alpha-amylase enzyme active site amino acids. This material is available as part of the online article from http://www.scielo.br/BJB ). The detailed parameters indicating the molecular docking affinities between the enzyme and possible binding molecules are shown (Table 2).
The interaction of amylopectin (PubChem ID - 439207) and alpha-amylase enzyme active site. The left figure is built by Chimera and the right by MOE software.
Molecular docking analysis results, indicating the ID ligands and scores of different parameters.
4. Discussion
Amylase are essential enzyme used in carbohydrate metabolism by the hydrolysis of glycosidic bonds. This enzyme is widely distributed in natural world (Saburi et al., 2015SABURI, W., RACHI-OTSUKA, H., HONDOH, H., OKUYAMA, M., MORI, H. and KIMURA, A., 2015. Structural elements responsible for the glucosidic linkage-selectivity of a glycoside hydrolase family 13 exo-glucosidase. FEBS Letters, vol. 589, no. 7, pp. 865-869. http://dx.doi.org/10.1016/j.febslet.2015.02.023. PMid:25728274.
http://dx.doi.org/10.1016/j.febslet.2015...
) such as in plant, animals, fungi and bacteria (Ribeiro et al., 2000RIBEIRO, J.M., ROWTON, E.D. and CHARLAB, R., 2000. Salivary amylase activity of the phlebotomine sand fly, Lutzomyia longipalpis. Insect Biochemistry and Molecular Biology, vol. 30, no. 4, pp. 271-277. http://dx.doi.org/10.1016/S0965-1748(99)00119-8. PMid:10727893.
http://dx.doi.org/10.1016/S0965-1748(99)...
; Hagihara et al., 2001HAGIHARA, H., IGARASHI, K., HAYASHI, Y., ENDO, K., IKAWA-KITAYAMA, K., OZAKI, K., KAWAI, S. and ITO, S.. 2001. Novel α-Amylase That Is Highly Resistant to Chelating Reagents and Chemical Oxidants from the Alkaliphilic Bacillus Isolate KSM-K38. Applied and Environmental Microbiology, vol. 67, no. 4, pp. 1744-1750. http://dx.doi.org/10.1128/AEM.67.4.1744-1750.2001. PMid:11282629.
http://dx.doi.org/10.1128/AEM.67.4.1744-...
). Amylase have a lot of industrial applications (Bassinello et al., 2002BASSINELLO, P.Z., CORDENUNSI, B.R. and LAJOLO, F.M., 2002. Amylolytic activity in fruits: comparison of different substrates and methods using banana as model. Journal of Agricultural and Food Chemistry, vol. 50, no. 21, pp. 5781-5786. http://dx.doi.org/10.1021/jf011370p. PMid:12358438.
http://dx.doi.org/10.1021/jf011370p...
) including starch and its derivative industries like, detergent, textile, bakery, fermentation, paper making, ethanol production, food and brewing industry (Schroder et al., 2015SCHRODER, C., BLANK, S. and ANTRANIKIAN, G., 2015. First glycoside hydrolase family 2 enzymes from Thermus antranikianii and Thermus brockianus with β-glucosidase activity. Frontiers in Bioengineering and Biotechnology, vol. 3, pp. 76. PMid:26090361.). The objective of the present study was recombinant production, purification and properties of ɑ - amylase from Geobacillus thermodenitrificans DSM-465. A wide range of genes coding for alpha amylase with different size and sequence has been reported in the literature which constitute the amylase pool from known sources (Ju et al., 2019JU, L., PAN, Z., ZHANG, H., LI, Q., LIANG, J., DENG, G., YU, M. and LONG, H., 2019. New insights into the origin and evolution of α-amylase genes in green plants. Scientific Reports, vol. 9, no. 1, pp. 4929. http://dx.doi.org/10.1038/s41598-019-41420-w. PMid:30894656.
http://dx.doi.org/10.1038/s41598-019-414...
). The gene coding for alpha amylase from G. thermodenitrificans DSM-465 was PCR amplified and about 1700 bp PCR product was found on agarose gel. The gene was ligated in to pTZ57R/T plasmid by T/A cloning kit method (Thermo Scientific. InsTAclone PCR Cloning Kit #K1214), the ligation mixture was used for the transformation of E. coli cells. The subsequent white colonies were screened for the presence of recombinant plasmid by restriction analysis. The restricted amylase gene was subcloned into pET21a (+) plasmid vector. The white colonies were reconfirmed by the plasmid isolation and restriction analysis. The enzyme was expressed in the culture medium containing 0.5mM IPTG. IPTG is most preferred substance for the induction of recombinant proteins under T7 promoter system. Different IPTG concentrations ranging from 0.1mM to 1mM have been optimized in different studies with different proteins (Katzke et al., 2010KATZKE, N., ARVANI, S., BERGMANN, R., CIRCOLONE, F., MARKERT, A., SVENSSON, V., JAEGER, K.E., HECK, A. and DREPPER, T., 2010. A novel T7 RNA polymerase dependent expression system for high-level protein production in the phototrophic bacterium Rhodobacter capsulatus. Protein Expression and Purification, vol. 69, no. 2, pp. 137-146. http://dx.doi.org/10.1016/j.pep.2009.08.008. PMid:19706327.
http://dx.doi.org/10.1016/j.pep.2009.08....
; Fazaeli et al., 2019FAZAELI, A., GOLESTANI, A., LAKZAEI, M., RASI VARAEI, S.S. and AMINIAN, M., 2019. Expression optimization, purification, and functional characterization of cholesterol oxidase from Chromobacterium sp. DS1. PLoS One, vol. 14, no. 2, pp. e0212217. http://dx.doi.org/10.1371/journal.pone.0212217. PMid:30759160.
http://dx.doi.org/10.1371/journal.pone.0...
). On SDS-PAGE the enzyme exhibited a molecular weight of 63 kDa (Figure 1). The molecular weight determined by SDS-PAGE was slightly lower than the theoretical molecular weight (65694.27 Da) of enzyme calculated from its primary structure by ProtParam software. Amylases with a range of molecular weights have been reported in the literature. Enzyme with molecular weight of 12.2 kDa from Geobacillus sp. (Febriani et al., 2019FEBRIANI, RAYYANA, ULYA, M., OESMAN, F., AKHMALOKA. and IQBALSYAH, T.M., 2019. Low molecular weight alkaline thermostable α-amylase from Geobacillus sp. nov. Heliyon, vol. 5, no. 7, pp. e02171. http://dx.doi.org/10.1016/j.heliyon.2019.e02171. PMid:31388592.
http://dx.doi.org/10.1016/j.heliyon.2019...
), 30 kDa from Trichoderma pseudokoningii (Abdulaal, 2018ABDULAAL, W.H., 2018. Purification and characterization of α-amylase from Trichoderma pseudokoningii. BMC Biochemistry, vol. 19, no. 1, pp. 4. http://dx.doi.org/10.1186/s12858-018-0094-8. PMid:29902965.
http://dx.doi.org/10.1186/s12858-018-009...
), 68 kDa from Bacillus subtilis (Trabelsi et al., 2019TRABELSI, S., BEN MABROUK, S., KRIAA, M., AMERI, R., SAHNOUN, M., MEZGHANI, M. and BEJAR, S., 2019. The optimized production, purification, characterization, and application in the bread making industry of three acid‐stable alpha‐amylases isoforms from a new isolated Bacillus subtilis strain US586. Journal of Food Biochemistry, vol. 43, no. 5, pp. e12826. http://dx.doi.org/10.1111/jfbc.12826. PMid:31353531.
http://dx.doi.org/10.1111/jfbc.12826...
), 73 kDa from Dociostaurus maroccanus (Rafiei et al., 2016RAFIEI, B., GHADAMYARI, M., IMANI, S., HOSSEININAVEH, V. and AHADIYAT, A., 2016. Purification and characterization of α-amylase in Moroccan locust, Dociostaurus maroccanus Thunberg (Orthoptera: Acrididae) and its inhibition by inhibitors from Phaseolus vulgaris L. Toxin Reviews, vol. 35, no. 3-4, pp. 90-97. http://dx.doi.org/10.1080/15569543.2016.1207190.
http://dx.doi.org/10.1080/15569543.2016....
), 45 kDa from Bacillus sonorensis (Vyas et al., 2019VYAS, G., SHARMA, N. and SHARMA, N., 2019. Purification and characterization of α-amylase from a novel thermoalkalophilic strain of Bacillus sonorensis GV2 isolated from mushroom compost. International Research Journal of Pure and Applied Chemistry, vol. 25, pp. 1-4. http://dx.doi.org/10.9734/irjpac/2019/v19i330111.
http://dx.doi.org/10.9734/irjpac/2019/v1...
) have been reported.
The enzyme being reported in the present report has shown optimum activity at pH 8, 700C. It was stable at a pH range from 5 to 10 and higher temperatures (Figure 2). Enzymes with optimum activity at pH 5 to 8 and temperature between 30 to 90°C have been reported from many sources (Baltas et al., 2016BALTAS, N., DINCER, B., EKINCI, A.P., KOLAYLI, S. and ADIGUZEL, A., 2016. Purification and characterization of extracellular α-amylase from a thermophilic Anoxybacillus thermarum A4 strain. Brazilian Archives of Biology and Technology, vol. 59, pp. e16160346. http://dx.doi.org/10.1590/1678-4324-2016160346.
http://dx.doi.org/10.1590/1678-4324-2016...
; Hu et al., 2019HU, Y., YU, D., WANG, Z., HOU, J., TYAGI, R., LIANG, Y. and HU, Y., 2019. Purification and characterization of a novel, highly potent fibrinolytic enzyme from Bacillus subtilis DC27 screened from Douchi, a traditional Chinese fermented soybean food. Scientific Reports, vol. 9, no. 1, pp. 9235. http://dx.doi.org/10.1038/s41598-019-45686-y. PMid:31239529.
http://dx.doi.org/10.1038/s41598-019-456...
; Naidu et al., 2019NAIDU, K., MASEKO, S., KRUGER, G. and LIN, J., 2019. Purification and characterization of α-amylase from Paenibacillus sp. D9 and Escherichia coli recombinants. Biocatalysis and Biotransformation, vol. 38, no. 1, pp. 24-34.; Fang et al., 2019FANG, W., XUE, S., DENG, P., ZHANG, X., WANG, X., XIAO, Y. and FANG, Z., 2019. AmyZ1: a novel α-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches. Biotechnology for Biofuels, vol. 12, no. 1, pp. 95. http://dx.doi.org/10.1186/s13068-019-1432-9. PMid:31044008.
http://dx.doi.org/10.1186/s13068-019-143...
; Al-Dhabi et al., 2020AL-DHABI, N.A., ESMAIL, G.A., GHILAN, A.K., ARASU, M.V., DURAIPANDIYAN, V. and PONMURUGAN, K., 2020. Isolation and purification of starch hydrolysing amylase from Streptomyces sp. Al-Dhabi-46 obtained from the Jazan region of Saudi Arabia with industrial applications. Journal of King Saud University-Science, vol. 32, no. 1, pp. 1226-1232. http://dx.doi.org/10.1016/j.jksus.2019.11.018.
http://dx.doi.org/10.1016/j.jksus.2019.1...
). The KM value of recombinant enzyme was 157.7µM and Vmax was found 333.3 micromoles per minute as determined by Lineweaver-burk plot and subsequent calculations. Amylases with different KM and Vmax values have been reported from different species. As for example, the enzyme from Trichoderma pseudokoningii has KM and Vmax of 0.74µM reducing sugar and 4mg of starch respectively (Abdulaal, 2018ABDULAAL, W.H., 2018. Purification and characterization of α-amylase from Trichoderma pseudokoningii. BMC Biochemistry, vol. 19, no. 1, pp. 4. http://dx.doi.org/10.1186/s12858-018-0094-8. PMid:29902965.
http://dx.doi.org/10.1186/s12858-018-009...
), these values were reported 37.3 mM and 19.8 U/mg for the enzyme isolated from Paenibacillus sp (Roth et al., 2019ROTH, C., MOROZ, O.V., TURKENBURG, J.P., BLAGOVA, E., WATERMAN, J., ARIZA, A., MING, L., TIANQI, S., ANDERSEN, C., DAVIES, G.J. and WILSON, K.S., 2019. structural and functional characterization of three novel fungal amylases with enhanced stability and pH tolerance. International Journal of Molecular Sciences, vol. 20, no. 19, pp. 4902. http://dx.doi.org/10.3390/ijms20194902. PMid:31623309.
http://dx.doi.org/10.3390/ijms20194902...
), 1347 μmol/mg/min and 3.46 mmol/ml for the amylase purified from Bacillus sonorensis (Vyas et al., 2019VYAS, G., SHARMA, N. and SHARMA, N., 2019. Purification and characterization of α-amylase from a novel thermoalkalophilic strain of Bacillus sonorensis GV2 isolated from mushroom compost. International Research Journal of Pure and Applied Chemistry, vol. 25, pp. 1-4. http://dx.doi.org/10.9734/irjpac/2019/v19i330111.
http://dx.doi.org/10.9734/irjpac/2019/v1...
) The enzyme was efficiently applied for the desizing of textile at high temperatures and it was able to remove up to 95% of starch from the cotton fabric. In silico studies have shown that the enzyme exists as a monomer in Geobacillus. Monomeric amylases have been reported in literature from various species (Febriani et al., 2019FEBRIANI, RAYYANA, ULYA, M., OESMAN, F., AKHMALOKA. and IQBALSYAH, T.M., 2019. Low molecular weight alkaline thermostable α-amylase from Geobacillus sp. nov. Heliyon, vol. 5, no. 7, pp. e02171. http://dx.doi.org/10.1016/j.heliyon.2019.e02171. PMid:31388592.
http://dx.doi.org/10.1016/j.heliyon.2019...
; Roth et al., 2019ROTH, C., MOROZ, O.V., TURKENBURG, J.P., BLAGOVA, E., WATERMAN, J., ARIZA, A., MING, L., TIANQI, S., ANDERSEN, C., DAVIES, G.J. and WILSON, K.S., 2019. structural and functional characterization of three novel fungal amylases with enhanced stability and pH tolerance. International Journal of Molecular Sciences, vol. 20, no. 19, pp. 4902. http://dx.doi.org/10.3390/ijms20194902. PMid:31623309.
http://dx.doi.org/10.3390/ijms20194902...
). The 3D model was built by Raptor-X software (Källberg et al., 2014KÄLLBERG, M., MARGARYAN, G., WANG, S., MA, J. and XU, J., 2014. RaptorX server: a resource for template-based protein structure modeling. In Protein structure prediction. New York, NY: Humana Press, pp. 17-27.) (Figure 3) and validated by Ramachandran plot (Laskowski et al., 2013LASKOWSKI, R.A., FURNHAM, N. and THORNTON, J.M., 2013. The Ramachandran plot and protein structure validation. In M. BANSAL, ed. Biomolecular forms and functions: a celebration of 50 years of the ramachandran map. Bangalore, India: World Sicentific, pp. 62-75. http://dx.doi.org/10.1142/9789814449144_0005.
http://dx.doi.org/10.1142/9789814449144_...
; Pražnikar et al., 2019PRAŽNIKAR, J., TOMIĆ, M. and TURK, D., 2019. Validation and quality assessment of macromolecular structures using complex network analysis. Scientific Reports, vol. 9, no. 1, pp. 1678. http://dx.doi.org/10.1038/s41598-019-38658-9. PMid:30737447.
http://dx.doi.org/10.1038/s41598-019-386...
) (Figure 4). The molecular docking studies have indicated a higher affinity of enzyme for starch, amylopectin and glycogen molecules. With higher energy changes, amylopectin (∆G -10.59 Kcal/mole) (Figure 5), Glycogen (∆G -10.41Kcal/mole) (Supplementary material
Supplementary Material
Supplementary material accompanies this paper
Figure S1. The interaction of glycogen molecule (PubChem ID - 439177) and alpha-amylase enzyme active site. Glycogen was the second most strongly binding molecule with amylase structure.
Figure S2. Molecular interaction of starch molecule (PubChem ID - 24836924) and alpha-amylase enzyme active site amino acids as determined by molecular docking analysis.
Figure S3. Docking based interaction of amylose molecule (PubChem ID - 53477771) and alpha-amylase enzyme active site amino acids.
Figure S4. Molecular docking results showing the interaction of maltose molecule (PubChem ID - 439341) and alpha-amylase enzyme active site amino acids.
This material is available as part of the online article from http://www.scielo.br/BJB
), and Starch (∆G -10.04 Kcal/mole) (Supplementary material
Supplementary Material
Supplementary material accompanies this paper
Figure S1. The interaction of glycogen molecule (PubChem ID - 439177) and alpha-amylase enzyme active site. Glycogen was the second most strongly binding molecule with amylase structure.
Figure S2. Molecular interaction of starch molecule (PubChem ID - 24836924) and alpha-amylase enzyme active site amino acids as determined by molecular docking analysis.
Figure S3. Docking based interaction of amylose molecule (PubChem ID - 53477771) and alpha-amylase enzyme active site amino acids.
Figure S4. Molecular docking results showing the interaction of maltose molecule (PubChem ID - 439341) and alpha-amylase enzyme active site amino acids.
This material is available as part of the online article from http://www.scielo.br/BJB
), indicating that starch, glycogen and amylopectin establish more stable interaction with the enzyme cavity. Maltose has the lowest binding affinity with the enzyme with estimated binding energy ∆G -2.8 Kcal/mole (Supplementary material
Supplementary Material
Supplementary material accompanies this paper
Figure S1. The interaction of glycogen molecule (PubChem ID - 439177) and alpha-amylase enzyme active site. Glycogen was the second most strongly binding molecule with amylase structure.
Figure S2. Molecular interaction of starch molecule (PubChem ID - 24836924) and alpha-amylase enzyme active site amino acids as determined by molecular docking analysis.
Figure S3. Docking based interaction of amylose molecule (PubChem ID - 53477771) and alpha-amylase enzyme active site amino acids.
Figure S4. Molecular docking results showing the interaction of maltose molecule (PubChem ID - 439341) and alpha-amylase enzyme active site amino acids.
This material is available as part of the online article from http://www.scielo.br/BJB
), (Table 2). Amylase with ability to degrade starch and glycogen molecules have been reported in the literature (Van der Maarel et al., 2002VAN DER MAAREL, M.J., VAN DER VEEN, B., UITDEHAAG, J.C., LEEMHUIS, H. and DIJKHUIZEN, L., 2002. Properties and applications of starch-converting enzymes of the α-amylase family. Journal of Biotechnology, vol. 94, no. 2, pp. 137-155. http://dx.doi.org/10.1016/S0168-1656(01)00407-2. PMid:11796168.
http://dx.doi.org/10.1016/S0168-1656(01)...
; Reddy et al., 2003REDDY, N., NIMMAGADDA, A. and RAO, K.S., 2003. An overview of the microbial α-amylase family. African Journal of Biotechnology, vol. 2, no. 12, pp. 645-648. http://dx.doi.org/10.5897/AJB2003.000-1119.
http://dx.doi.org/10.5897/AJB2003.000-11...
; Yan and Wu, 2017YAN, S. and WU, G., 2017. Bottleneck in secretion of α-amylase in Bacillus subtilis. Microbial Cell Factories, vol. 16, no. 1, pp. 124. http://dx.doi.org/10.1186/s12934-017-0738-1. PMid:28724440.
http://dx.doi.org/10.1186/s12934-017-073...
). According to our molecular docking studies, Asp232, Glu274, Arg448, Glu385, Asp34, Asn276, and Arg175 are the main residues interacting with the protein partner molecules. These findings suggest an active site for the enzyme. Asp and Glu residues have been reported as the active site amino acids of amylase from human pancreas and other species (Brayer et al., 2000BRAYER, G.D., SIDHU, G., MAURUS, R., RYDBERG, E.H., BRAUN, C., WANG, Y., NGUYEN, N.T., OVERALL, C.M. and WITHERS, S.G., 2000. Subsite mapping of the human pancreatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry, vol. 39, no. 16, pp. 4778-4791. http://dx.doi.org/10.1021/bi9921182. PMid:10769135.
http://dx.doi.org/10.1021/bi9921182...
). In conclusion, α - amylase from Geobacillus thermodenitrificans DSM-465 has been produced in E. coli by recombinant DNA technology, purified and characterized for its kinetic properties. The enzyme is stable at a pH between 6 to 10 and 55 to 80°C temperature which makes it a good candidate for industrial applications. In silico model and molecular docking studies have been used to predict the 3D structure and active site residues of subject. The enzyme has shown high efficacy to desize the cotton fabric indicating its potential application in the industry. The report will add to the body of knowledge about amylase from this species in particular and about amylases in general.
Supplementary Material
Supplementary material accompanies this paper
Figure S1. The interaction of glycogen molecule (PubChem ID - 439177) and alpha-amylase enzyme active site. Glycogen was the second most strongly binding molecule with amylase structure.
Figure S2. Molecular interaction of starch molecule (PubChem ID - 24836924) and alpha-amylase enzyme active site amino acids as determined by molecular docking analysis.
Figure S3. Docking based interaction of amylose molecule (PubChem ID - 53477771) and alpha-amylase enzyme active site amino acids.
Figure S4. Molecular docking results showing the interaction of maltose molecule (PubChem ID - 439341) and alpha-amylase enzyme active site amino acids.
This material is available as part of the online article from http://www.scielo.br/BJB
Acknowledgements
The authors are thankful to DSR at King Abdulaziz University Jeddah for supporting this research.
References
- ABDULAAL, W.H., 2018. Purification and characterization of α-amylase from Trichoderma pseudokoningii. BMC Biochemistry, vol. 19, no. 1, pp. 4. http://dx.doi.org/10.1186/s12858-018-0094-8 PMid:29902965.
» http://dx.doi.org/10.1186/s12858-018-0094-8 - AHMED, S., ZIA, A., MEHMOOD, S.A., PANHWAR, W.A., KHAN, W., SHAH, M., ULLAH, I., 2021. Change in malate dehydrogenase and alpha amylase activities in Rubus fruticosus and Valeriana jatamansi treated granary weevil, Sitophilus granarius Brazilian Journal of Biology = Revista Brasileira de Biologia, vol. 81, no. 2, pp. 387-391. http://dx.doi.org/10.1590/1519-6984.226952.
» https://doi.org/10.1590/1519-6984.226952 - AL-DHABI, N.A., ESMAIL, G.A., GHILAN, A.K., ARASU, M.V., DURAIPANDIYAN, V. and PONMURUGAN, K., 2020. Isolation and purification of starch hydrolysing amylase from Streptomyces sp. Al-Dhabi-46 obtained from the Jazan region of Saudi Arabia with industrial applications. Journal of King Saud University-Science, vol. 32, no. 1, pp. 1226-1232. http://dx.doi.org/10.1016/j.jksus.2019.11.018
» http://dx.doi.org/10.1016/j.jksus.2019.11.018 - ALLAN, S., TORBENVEDEL, B. and HENRICK, B., 1997. Recombinant alpha amylase mutants and their use in textile desizing starch liquification and washing. PTC International Patent Application, vol. 12, no. 5, pp. 205-210.
- ASGHER, M., ASAD, M.J., RAHMAN, S.U. and LEGGE, R.L., 2007. A thermostable α-amylase from a moderately thermophilic Bacillus subtilis strain for starch processing. Journal of Food Engineering, vol. 79, no. 3, pp. 950-955. http://dx.doi.org/10.1016/j.jfoodeng.2005.12.053
» http://dx.doi.org/10.1016/j.jfoodeng.2005.12.053 - BALTAS, N., DINCER, B., EKINCI, A.P., KOLAYLI, S. and ADIGUZEL, A., 2016. Purification and characterization of extracellular α-amylase from a thermophilic Anoxybacillus thermarum A4 strain. Brazilian Archives of Biology and Technology, vol. 59, pp. e16160346. http://dx.doi.org/10.1590/1678-4324-2016160346
» http://dx.doi.org/10.1590/1678-4324-2016160346 - BASSINELLO, P.Z., CORDENUNSI, B.R. and LAJOLO, F.M., 2002. Amylolytic activity in fruits: comparison of different substrates and methods using banana as model. Journal of Agricultural and Food Chemistry, vol. 50, no. 21, pp. 5781-5786. http://dx.doi.org/10.1021/jf011370p PMid:12358438.
» http://dx.doi.org/10.1021/jf011370p - BRADFORD, M.M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, vol. 72, pp. 248-254. http://dx.doi.org/10.1016/0003-2697(76)90527-3 PMid:942051.
» http://dx.doi.org/10.1016/0003-2697(76)90527-3 - BRAYER, G.D., SIDHU, G., MAURUS, R., RYDBERG, E.H., BRAUN, C., WANG, Y., NGUYEN, N.T., OVERALL, C.M. and WITHERS, S.G., 2000. Subsite mapping of the human pancreatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry, vol. 39, no. 16, pp. 4778-4791. http://dx.doi.org/10.1021/bi9921182 PMid:10769135.
» http://dx.doi.org/10.1021/bi9921182 - CELNIKER, G., NIMROD, G., ASHKENAZY, H., GLASER, F., MARTZ, E., MAYROSE, I., PUPKO, T. and BEN‐TAL, N., 2013. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Israel Journal of Chemistry, vol. 53, no. 3-4, pp. 199-206. http://dx.doi.org/10.1002/ijch.201200096
» http://dx.doi.org/10.1002/ijch.201200096 - FANG, W., XUE, S., DENG, P., ZHANG, X., WANG, X., XIAO, Y. and FANG, Z., 2019. AmyZ1: a novel α-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches. Biotechnology for Biofuels, vol. 12, no. 1, pp. 95. http://dx.doi.org/10.1186/s13068-019-1432-9 PMid:31044008.
» http://dx.doi.org/10.1186/s13068-019-1432-9 - FAZAELI, A., GOLESTANI, A., LAKZAEI, M., RASI VARAEI, S.S. and AMINIAN, M., 2019. Expression optimization, purification, and functional characterization of cholesterol oxidase from Chromobacterium sp. DS1. PLoS One, vol. 14, no. 2, pp. e0212217. http://dx.doi.org/10.1371/journal.pone.0212217 PMid:30759160.
» http://dx.doi.org/10.1371/journal.pone.0212217 - FEBRIANI, RAYYANA, ULYA, M., OESMAN, F., AKHMALOKA. and IQBALSYAH, T.M., 2019. Low molecular weight alkaline thermostable α-amylase from Geobacillus sp. nov. Heliyon, vol. 5, no. 7, pp. e02171. http://dx.doi.org/10.1016/j.heliyon.2019.e02171 PMid:31388592.
» http://dx.doi.org/10.1016/j.heliyon.2019.e02171 - FURNHAM, N., DORÉ, A.S., CHIRGADZE, D.Y., DE BAKKER, P.I., DEPRISTO, M.A. and BLUNDELL, T.L., 2006. Knowledge-based real-space explorations for low-resolution structure determination. Structure (London, England : 1993), vol. 14, no. 8, pp. 1313-1320. http://dx.doi.org/10.1016/j.str.2006.06.014 PMid:16905105.
» http://dx.doi.org/10.1016/j.str.2006.06.014 - GARG, G., SINGH, A., KAUR, A., SINGH, R., KAUR, J., MAHAJAN, R., 2016. Microbial pectinases: an ecofriendly tool of nature for industries. 3 Biotech, vol. 6, no. 1, pp. 47.
- GHASEMI, A., GHAFOURIAN, S., VAFAEI, S., MOHEBI, R., FARZI, M., TAHERIKALANI, M. and SADEGHIFARD, N., 2015. Cloning, Expression, and Purification of Hyperthermophile α-Amylase from Pyrococcus woesei. Osong Public Health and Research Perspectives, vol. 6, no. 6, pp. 336-340. http://dx.doi.org/10.1016/j.phrp.2015.10.003 PMid:26835242.
» http://dx.doi.org/10.1016/j.phrp.2015.10.003 - GUPTA, R., GIGRAS, P., MOHAPATRA, H., GOSWAMI, V.K. and CHAUHAN, B., 2003. Microbial α-amylases: a biotechnological perspective. Process Biochemistry, vol. 38, no. 11, pp. 1599-1616. http://dx.doi.org/10.1016/S0032-9592(03)00053-0
» http://dx.doi.org/10.1016/S0032-9592(03)00053-0 - HAGIHARA, H., IGARASHI, K., HAYASHI, Y., ENDO, K., IKAWA-KITAYAMA, K., OZAKI, K., KAWAI, S. and ITO, S.. 2001. Novel α-Amylase That Is Highly Resistant to Chelating Reagents and Chemical Oxidants from the Alkaliphilic Bacillus Isolate KSM-K38. Applied and Environmental Microbiology, vol. 67, no. 4, pp. 1744-1750. http://dx.doi.org/10.1128/AEM.67.4.1744-1750.2001 PMid:11282629.
» http://dx.doi.org/10.1128/AEM.67.4.1744-1750.2001 - HU, Y., YU, D., WANG, Z., HOU, J., TYAGI, R., LIANG, Y. and HU, Y., 2019. Purification and characterization of a novel, highly potent fibrinolytic enzyme from Bacillus subtilis DC27 screened from Douchi, a traditional Chinese fermented soybean food. Scientific Reports, vol. 9, no. 1, pp. 9235. http://dx.doi.org/10.1038/s41598-019-45686-y PMid:31239529.
» http://dx.doi.org/10.1038/s41598-019-45686-y - JU, L., PAN, Z., ZHANG, H., LI, Q., LIANG, J., DENG, G., YU, M. and LONG, H., 2019. New insights into the origin and evolution of α-amylase genes in green plants. Scientific Reports, vol. 9, no. 1, pp. 4929. http://dx.doi.org/10.1038/s41598-019-41420-w PMid:30894656.
» http://dx.doi.org/10.1038/s41598-019-41420-w - KÄLLBERG, M., MARGARYAN, G., WANG, S., MA, J. and XU, J., 2014. RaptorX server: a resource for template-based protein structure modeling. In Protein structure prediction New York, NY: Humana Press, pp. 17-27.
- KÄLLBERG, M., WANG, H., WANG, S., PENG, J., WANG, Z., LU, H. and XU, J., 2012. Template-based protein structure modeling using the RaptorX web server. Nature Protocols, vol. 7, no. 8, pp. 1511-1522. http://dx.doi.org/10.1038/nprot.2012.085 PMid:22814390.
» http://dx.doi.org/10.1038/nprot.2012.085 - KATZKE, N., ARVANI, S., BERGMANN, R., CIRCOLONE, F., MARKERT, A., SVENSSON, V., JAEGER, K.E., HECK, A. and DREPPER, T., 2010. A novel T7 RNA polymerase dependent expression system for high-level protein production in the phototrophic bacterium Rhodobacter capsulatus. Protein Expression and Purification, vol. 69, no. 2, pp. 137-146. http://dx.doi.org/10.1016/j.pep.2009.08.008 PMid:19706327.
» http://dx.doi.org/10.1016/j.pep.2009.08.008 - KUMAR, S., 2015. Role of enzymes in fruit juice processing and its quality enhancement. Health, vol. 6, no. 6, pp. 114-124.
- LASKOWSKI, R.A., FURNHAM, N. and THORNTON, J.M., 2013. The Ramachandran plot and protein structure validation. In M. BANSAL, ed. Biomolecular forms and functions: a celebration of 50 years of the ramachandran map Bangalore, India: World Sicentific, pp. 62-75. http://dx.doi.org/10.1142/9789814449144_0005
» http://dx.doi.org/10.1142/9789814449144_0005 - MEHTA, D. and SATYANARAYANA, T., 2016. Bacterial and archaeal α-amylases: diversity and amelioration of the desirable characteristics for industrial applications. Frontiers in Microbiology, vol. 7, pp. 1129. http://dx.doi.org/10.3389/fmicb.2016.01129 PMid:27516755.
» http://dx.doi.org/10.3389/fmicb.2016.01129 - NAIDU, K., MASEKO, S., KRUGER, G. and LIN, J., 2019. Purification and characterization of α-amylase from Paenibacillus sp. D9 and Escherichia coli recombinants. Biocatalysis and Biotransformation, vol. 38, no. 1, pp. 24-34.
- PANDEY, A., NIGAM, P., SOCCOL, C.R., SOCCOL, V.T., SINGH, D. and MOHAN, R., 2000. Advances in microbial amylases. Biotechnology and Applied Biochemistry, vol. 31, no. 2, pp. 135-152. http://dx.doi.org/10.1042/BA19990073 PMid:10744959.
» http://dx.doi.org/10.1042/BA19990073 - PETTERSEN, E.F., GODDARD, T.D., HUANG, C.C., COUCH, G.S., GREENBLATT, D.M., MENG, E.C. and FERRIN, T.E., 2004. UCSF Chimera: a visualization system for exploratory research and analysis. Journal of Computational Chemistry, vol. 25, no. 13, pp. 1605-1612. http://dx.doi.org/10.1002/jcc.20084 PMid:15264254.
» http://dx.doi.org/10.1002/jcc.20084 - PRAŽNIKAR, J., TOMIĆ, M. and TURK, D., 2019. Validation and quality assessment of macromolecular structures using complex network analysis. Scientific Reports, vol. 9, no. 1, pp. 1678. http://dx.doi.org/10.1038/s41598-019-38658-9 PMid:30737447.
» http://dx.doi.org/10.1038/s41598-019-38658-9 - RAFIEI, B., GHADAMYARI, M., IMANI, S., HOSSEININAVEH, V. and AHADIYAT, A., 2016. Purification and characterization of α-amylase in Moroccan locust, Dociostaurus maroccanus Thunberg (Orthoptera: Acrididae) and its inhibition by inhibitors from Phaseolus vulgaris L. Toxin Reviews, vol. 35, no. 3-4, pp. 90-97. http://dx.doi.org/10.1080/15569543.2016.1207190
» http://dx.doi.org/10.1080/15569543.2016.1207190 - RAO, M.B., TANKSALE, A.M., GHATGE, M.S. and DESHPANDE, V.V., 1998. Molecular and biotechnological aspects of microbial proteases. Microbiology and Molecular Biology Reviews, vol. 62, no. 3, pp. 597-635. http://dx.doi.org/10.1128/MMBR.62.3.597-635.1998 PMid:9729602.
» http://dx.doi.org/10.1128/MMBR.62.3.597-635.1998 - REDDY, N., NIMMAGADDA, A. and RAO, K.S., 2003. An overview of the microbial α-amylase family. African Journal of Biotechnology, vol. 2, no. 12, pp. 645-648. http://dx.doi.org/10.5897/AJB2003.000-1119
» http://dx.doi.org/10.5897/AJB2003.000-1119 - RIBEIRO, J.M., ROWTON, E.D. and CHARLAB, R., 2000. Salivary amylase activity of the phlebotomine sand fly, Lutzomyia longipalpis. Insect Biochemistry and Molecular Biology, vol. 30, no. 4, pp. 271-277. http://dx.doi.org/10.1016/S0965-1748(99)00119-8 PMid:10727893.
» http://dx.doi.org/10.1016/S0965-1748(99)00119-8 - ROTH, C., MOROZ, O.V., TURKENBURG, J.P., BLAGOVA, E., WATERMAN, J., ARIZA, A., MING, L., TIANQI, S., ANDERSEN, C., DAVIES, G.J. and WILSON, K.S., 2019. structural and functional characterization of three novel fungal amylases with enhanced stability and pH tolerance. International Journal of Molecular Sciences, vol. 20, no. 19, pp. 4902. http://dx.doi.org/10.3390/ijms20194902 PMid:31623309.
» http://dx.doi.org/10.3390/ijms20194902 - SABURI, W., RACHI-OTSUKA, H., HONDOH, H., OKUYAMA, M., MORI, H. and KIMURA, A., 2015. Structural elements responsible for the glucosidic linkage-selectivity of a glycoside hydrolase family 13 exo-glucosidase. FEBS Letters, vol. 589, no. 7, pp. 865-869. http://dx.doi.org/10.1016/j.febslet.2015.02.023 PMid:25728274.
» http://dx.doi.org/10.1016/j.febslet.2015.02.023 - SAHA, P., KHAN, M.F. and PATRA, S., 2018. Truncated α-amylase: an improved candidate for textile processing. Preparative Biochemistry & Biotechnology, vol. 48, no. 7, pp. 635-645. http://dx.doi.org/10.1080/10826068.2018.1479863 PMid:29902115.
» http://dx.doi.org/10.1080/10826068.2018.1479863 - SAINI, R., SAINI, H.S. and DAHIYA, A., 2017. Amylases: characteristics and industrial applications. Journal of Pharmacognosy and Phytochemistry, vol. 6, no. 4, pp. 1865-1871.
- SAPUNDZHI, F.I. and DZIMBOVA, T.A., 2018. Computer modelling of the CB1 receptor by molecular operating environment. Bulgarian Chemical Communications, vol. 50. Special Issue B, pp. 15-19.
- SARIAN, F.D., JANEČEK, Š., PIJNING, T., NURACHMAN, Z., RADJASA, O.K., DIJKHUIZEN, L., NATALIA, D. and VAN DER MAAREL, M.J., 2017. A new group of glycoside hydrolase family 13 α-amylases with an aberrant catalytic triad. Scientific Reports, vol. 71, pp. 1-10.
- SCHRODER, C., BLANK, S. and ANTRANIKIAN, G., 2015. First glycoside hydrolase family 2 enzymes from Thermus antranikianii and Thermus brockianus with β-glucosidase activity. Frontiers in Bioengineering and Biotechnology, vol. 3, pp. 76. PMid:26090361.
- SIDHU, G., SHARMA, P., CHAKRABARTI, T. and GUPTA, J., 1997. Strain improvement for the production of a thermostable α-amylase. Enzyme and Microbial Technology, vol. 21, no. 7, pp. 525-530. http://dx.doi.org/10.1016/S0141-0229(97)00055-0
» http://dx.doi.org/10.1016/S0141-0229(97)00055-0 - SIVARAMAKRISHNAN, S., GANGADHARAN, D., NAMPOOTHIRI, K.M., SOCCOL, C.R. and PANDEY, A., 2006. Alpha - Amylases from microbial sources–an overview on recent developments. Food Technology and Biotechnology, vol. 44, no. 2, pp. 173-184.
- SUNDARRAM, A. and MURTHY, T.P.K., 2014. α-amylase production and applications: a review. Journal of Applied & Environmental Microbiology, vol. 2, no. 4, pp. 166-175.
- TRABELSI, S., BEN MABROUK, S., KRIAA, M., AMERI, R., SAHNOUN, M., MEZGHANI, M. and BEJAR, S., 2019. The optimized production, purification, characterization, and application in the bread making industry of three acid‐stable alpha‐amylases isoforms from a new isolated Bacillus subtilis strain US586. Journal of Food Biochemistry, vol. 43, no. 5, pp. e12826. http://dx.doi.org/10.1111/jfbc.12826 PMid:31353531.
» http://dx.doi.org/10.1111/jfbc.12826 - VAN DER MAAREL, M.J., VAN DER VEEN, B., UITDEHAAG, J.C., LEEMHUIS, H. and DIJKHUIZEN, L., 2002. Properties and applications of starch-converting enzymes of the α-amylase family. Journal of Biotechnology, vol. 94, no. 2, pp. 137-155. http://dx.doi.org/10.1016/S0168-1656(01)00407-2 PMid:11796168.
» http://dx.doi.org/10.1016/S0168-1656(01)00407-2 - VYAS, G., SHARMA, N. and SHARMA, N., 2019. Purification and characterization of α-amylase from a novel thermoalkalophilic strain of Bacillus sonorensis GV2 isolated from mushroom compost. International Research Journal of Pure and Applied Chemistry, vol. 25, pp. 1-4. http://dx.doi.org/10.9734/irjpac/2019/v19i330111
» http://dx.doi.org/10.9734/irjpac/2019/v19i330111 - WANG S, LI W, LIU S, XU J., 2016. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Research, vol. 44, no. W1, pp. W430-W435.
- WANG, G., LUO, M., LIN, J., LIN, Y., YAN, R., STREIT, W.R. and YE, X., 2019. A new extremely halophilic, calcium-independent and surfactant-resistant alpha-amylase from Alkalibacterium sp SL3. Journal of Microbiology and Biotechnology, vol. 29, no. 5, pp. 765-775. http://dx.doi.org/10.4014/jmb.1901.01038 PMid:30982319.
» http://dx.doi.org/10.4014/jmb.1901.01038 - WATERHOUSE, A., BERTONI, M., BIENERT, S., STUDER, G., TAURIELLO, G., GUMIENNY, R., HEER, F.T., DE BEER, T.A.P., REMPFER, C., BORDOLI, L., LEPORE, R. and SCHWEDE, T., 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research, vol. 46, no. W1, pp. W296-W303. http://dx.doi.org/10.1093/nar/gky427 PMid:29788355.
» http://dx.doi.org/10.1093/nar/gky427 - YAN, S. and WU, G., 2017. Bottleneck in secretion of α-amylase in Bacillus subtilis. Microbial Cell Factories, vol. 16, no. 1, pp. 124. http://dx.doi.org/10.1186/s12934-017-0738-1 PMid:28724440.
» http://dx.doi.org/10.1186/s12934-017-0738-1 - YANG, G., YAO, H., MOZZICAFREDDO, M., BALLARINI, P., PUCCIARELLI, S. and MICELI, C., 2017. Rational engineering of a cold-adapted α-amylase from the Antarctic ciliate Euplotes focardii for simultaneous improvement of thermostability and catalytic activity. Applied and Environmental Microbiology, vol. 83, no. 13, pp. e00449-17. http://dx.doi.org/10.1128/AEM.00449-17 PMid:28455329.
» http://dx.doi.org/10.1128/AEM.00449-17 - YANG, J. and ZHANG, Y., 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Research, vol. 43, no. W1, pp. W174-W181. http://dx.doi.org/10.1093/nar/gkv342 PMid:25883148.
» http://dx.doi.org/10.1093/nar/gkv342 - ZAFAR, A., AFTAB, M.N., IQBAL, I., DIN, Z. and SALEEM, M.A., 2019. Pilot-scale production of a highly thermostable α-amylase enzyme from Thermotoga petrophila cloned into E. coli and its application as a desizer in textile industry. RSC Advances, vol. 9, no. 2, pp. 984-992. http://dx.doi.org/10.1039/C8RA06554C
» http://dx.doi.org/10.1039/C8RA06554C - ZEIGLER, D. R., 2014. The Geobacillus paradox: why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology, vol. 160, pp. 1-11. https://doi.org/10.1099/mic.0.071696-0
» https://doi.org/10.1099/mic.0.071696-0
Publication Dates
-
Publication in this collection
04 June 2021 -
Date of issue
2022
History
-
Received
11 June 2020 -
Accepted
29 Oct 2020