Abstract
Production of transgenic plants with desired agronomic and horticultural traits has gained great importance to fulfill demands of the growing population. Genetic transformation is also a fundamental step to study basics of plant sciences. Different transformation protocols have been developed and used which are reliable and efficient. These protocols used antibiotic or herbicide resistance genes incorporated along with gene of interest to identify transformed plants from non-transformed ones. These marker genes may pose a threat to human and environment. Use of visual markers enables direct and easier observation of transformed plants with more precision. In current study a gene cassette with ‘pigment production hydroxylase (PPH) gene under fiber specific promoter (GhSCFP) and downstream Nos-terminator was designed. After checking the structural and functional efficiency of codon optimized gene using bioinformatics tools, the cassette was sent for chemical synthesis from commercial source. The pigment gene cassette (PPH_CEMB), cloned in pCAMBIA-1301, was transformed into Agrobacterium through electroporation. Agrobacterium-mediated floral dip method was used to transform Camelina sativa inflorescence. After seed setting a total of 600 seed were observed for change in color and out of these, 19 seeds developed a reddish-brown coloration, while the remaining 581 seeds remained yellow. The transformation efficiency calculated on basis of color change was 1.0%. PCR analysis of leaves obtained after sowing reddish seeds confirmed the transformation of pigment production gene, while no PCR amplification was observed in leaves of plants from wild type seeds. From the results it is evident that Agrobacterium-mediated transformation of C. sativa inflorescence is very efficient and environment friendly technique not only for detection of transformed plants but also to study basic cellular processes.
Keywords:
Pigment Production Hydroxylase (PPH); biomarker; transgenic plants; Camelina sativa; floral dip method
Resumo
A produção de plantas transgênicas com características agronômicas e hortícolas desejadas vem ganhando grande importância por atender às demandas da crescente população. A transformação genética também é um passo fundamental para estudar os fundamentos das ciências das plantas. Foram desenvolvidos e utilizados diferentes protocolos de transformação, que são confiáveis e eficientes. Esses protocolos usaram genes de resistência a antibióticos ou herbicidas incorporados ao gene de interesse para identificar plantas transformadas e não transformadas. Esses genes marcadores podem representar uma ameaça ao ser humano e ao meio ambiente. O uso de marcadores visuais permite a observação direta e fácil de plantas transformadas com mais precisão. No estudo atual, foi projetado um cassete de genes com o gene da hidroxilase de produção de pigmentos (PPH) sob promotor específico de fibra (GhSCFP) e terminador NOS a jusante. Após verificar a eficiência estrutural e funcional do gene otimizado por códons usando ferramentas de bioinformática, o cassete foi enviado para síntese química a partir de fonte comercial. O cassete do gene do pigmento (PPH_CEMB), clonado em pCAMBIA-1301, foi transformado em Agrobacterium por eletroporação. O método de imersão floral mediado por Agrobacterium foi usado para transformar a inflorescência de Camelina sativa. Após a formação de sementes, foi observado um total de 600 sementes com mudança de cor, das quais 19 desenvolveram uma coloração marrom-avermelhada, enquanto as 581 restantes permaneceram amarelas. A eficiência de transformação calculada com base na mudança de cor foi de 1%. A análise de PCR das folhas obtidas após a semeadura de sementes avermelhadas confirmou a transformação do gene produtor de pigmentos, enquanto não foi observada amplificação por PCR em folhas de plantas de sementes do tipo selvagem. A partir dos resultados, é evidente que a transformação da inflorescência de C. sativa mediada por Agrobacterium é uma técnica muito eficiente e favorável ao ambiente não só para a detecção de plantas transformadas, mas também para estudar processos celulares básicos.
Palavras-chave:
Produção de Pigmentos Hidroxilase (PPH); biomarcador; plantas transgênicas; Camelina sativa; método floral dip
1. Introduction
Use of antibiotic marker genes is a common technique in selection of desired bacteria on the culture media. This concept is being utilized in genetically modified plants for selection of transformants. Concerns of the use of antibiotic resistance genes have been raised due to horizontal gene transfer (HGT) from transgenic plants to microbes (Nielsen et al., 1998NIELSEN, K.M., BONES, A.M., SMALLA, K. and VAN ELSAS, J.D., 1998. Horizontal gene transfer from transgenic plants to terrestrial bacteria–a rare event? FEMS Microbiology Reviews, vol. 22, no. 2, pp. 79-103. http://dx.doi.org/10.1016/S0168-6445(98)00009-6. PMid:9729765.
http://dx.doi.org/10.1016/S0168-6445(98)...
; Traavik and Ching, 2009TRAAVIK, T., and CHING, L.L., 2009. Biosafety first: holistic approaches to risk and uncertainty in genetic engineering and genetically modified organisms. Malaysia: Third World Network and GenØk.). This leads to increase in level of resistance towards antibiotics in microbes. This may pose a risk to human or animal health by compromising the therapeutic value of antibiotics for treatment of pathogenic micro-organisms (Pontiroli et al., 2007PONTIROLI, A., SIMONET, P., FROSTEGARD, A., VOGEL, T.M. and MONIER, J.-M., 2007. Fate of transgenic plant DNA in the environment. Environmental Biosafety Research, vol. 6, no. 1-2, pp. 15-35. http://dx.doi.org/10.1051/ebr:2007037. PMid:17961478.
http://dx.doi.org/10.1051/ebr:2007037...
). Transformation of the pigment genes in plant seeds has an advantage of elimination of antibiotic selection drug and screening of putative transformants by visual screening. Use of a visual or chromogenic marker is generally regarded as safe in production of transgenic plants where it is not desirable to have an antibiotic selectable marker. Antibiotic selectable marker genes often remain in the engineered plants adding time and cost to assess for safety and environmental impact (Van den Eede et al., 2004VAN DEN EEDE, G., AARTS, H., BUHK, H.-J., CORTHIER, G., FLINT, H.J., HAMMES, W., JACOBSEN, B., MIDTVEDT, T., VAN DER VOSSEN, J., VON WRIGHT, A., WACKERNAGEL, W. and WILCKS, A., 2004. The relevance of gene transfer to the safety of food and feed derived from genetically modified (GM) plants. Food and Chemical Toxicology, vol. 42, no. 7, pp. 1127-1156. http://dx.doi.org/10.1016/j.fct.2004.02.001. PMid:15123384.
http://dx.doi.org/10.1016/j.fct.2004.02....
; Costa-Font et al., 2008COSTA-FONT, M., GIL, J.M. and TRAILL, W.B., 2008. Consumer acceptance, valuation of and attitudes towards genetically modified food: review and implications for food policy. Food Policy, vol. 33, no. 2, pp. 99-111. http://dx.doi.org/10.1016/j.foodpol.2007.07.002.
http://dx.doi.org/10.1016/j.foodpol.2007...
).
Shoot apex method is a rapid genotype independent method of transformation and regeneration of cotton from shoots (embryos) isolated from germinating seedlings (Gould and Magallanes-Cedeno, 1998GOULD, J.H. and MAGALLANES-CEDENO, M., 1998. Adaptation of cotton shoot apex culture to Agrobacterium-mediated transformation. Plant Molecular Biology Reporter, vol. 16, no. 3, pp. 283-283. http://dx.doi.org/10.1023/A:1007438104369.
http://dx.doi.org/10.1023/A:100743810436...
; Rao et al., 2011RAO, A.Q., IRFAN, M., SALEEM, Z., NASIR, I.A., RIAZUDDIN, S. and HUSNAIN, T., 2011. Overexpression of the phytochrome B gene from Arabidopsis thaliana increases plant growth and yield of cotton (Gossypium hirsutum). Journal of Zhejiang University. Science. B., vol. 12, no. 4, pp. 326-334. http://dx.doi.org/10.1631/jzus.B1000168. PMid:21462389.
http://dx.doi.org/10.1631/jzus.B1000168...
). Isolated embryos are inoculated with Agrobacterium tumefaciens harboring pigment gene cassette, subjected to a mild antibiotic selection. These embryo re-generated shoots and roots in vitro. Rooted shoots can be obtained within 6–10 weeks of isolation and inoculation depending on the cotton cultivar (Gould and Magallanes-Cedeno, 1998GOULD, J.H. and MAGALLANES-CEDENO, M., 1998. Adaptation of cotton shoot apex culture to Agrobacterium-mediated transformation. Plant Molecular Biology Reporter, vol. 16, no. 3, pp. 283-283. http://dx.doi.org/10.1023/A:1007438104369.
http://dx.doi.org/10.1023/A:100743810436...
). Floral dip method of transformation is usually used in experiments which require quick expression in seeds. This method is usually used to transform seeds of a model plant C. sativa. Camelina inflorescence is dipped in a transformation solution having gene construct in A. tumefaciens strain LBA-4404 and applied the vacuum for five minutes. The plants are then shifted to growth room. The seeds are obtained after 8 weeks of transformation treatment (Schmidt and Pendarvis, 2017SCHMIDT, M.A. and PENDARVIS, K., 2017. Proteome rebalancing in transgenic Camelina occurs within the enlarged proteome induced by β-carotene accumulation and storage protein suppression. Transgenic Research, vol. 26, no. 2, pp. 171-186. http://dx.doi.org/10.1007/s11248-016-9992-y. PMid:27771868.
http://dx.doi.org/10.1007/s11248-016-999...
).
In the past Beyer et al. (2002)BEYER, P., AL-BABILI, S., YE, X., LUCCA, P., SCHAUB, P., WELSCH, R. and POTRYKUS, I., 2002. Golden rice: introducing the β-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency. The Journal of Nutrition, vol. 132, no. 3, pp. 506S-510S. http://dx.doi.org/10.1093/jn/132.3.506S. PMid:11880581.
http://dx.doi.org/10.1093/jn/132.3.506S...
transformed β-carotene pigment biosynthetic pathways into rice plant. They used phytoene synthase (psy) and lycopene β-cyclase (β-lcy) under endosperm specific promoter (glutelin promoter) and phytoene desaturase gene (crtI) from under 35S promoter (constitutive promoter). The phytoene synthase (psy) and lycopene β-cyclase (β-lcy) genes were taken from a wild daffodil plant (Narcissus pseudonarcissus) while Phytoene desaturase (crtI) gene was taken from a gram-negative bacterium (Erwinia uredovora). This combination of genes is sufficient for bio-synthesis of β-carotene pigment. Phenotypic analysis of T0 generation revealed presence of yellow colored rice containing β-carotene (Beyer et al., 2002BEYER, P., AL-BABILI, S., YE, X., LUCCA, P., SCHAUB, P., WELSCH, R. and POTRYKUS, I., 2002. Golden rice: introducing the β-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency. The Journal of Nutrition, vol. 132, no. 3, pp. 506S-510S. http://dx.doi.org/10.1093/jn/132.3.506S. PMid:11880581.
http://dx.doi.org/10.1093/jn/132.3.506S...
). Another attempt was made to transform melanin pigment pathway into the cotton plant. The Streptomyces antibiotics derived genes TyrA and ORF438 are responsible for the synthesis of melanin pigment. Variant of these genes was designed by codon optimization according to the nucleotide preference of cotton and resultant versions of these genes were given names dTyrA and dORF438. These genes were then chemically synthesized from a commercial source and cloned in a plant expression vector pBin under fiber specific promoter LTP3. The successful integration of these genes was confirmed through PCR analysis and melanin pigment was observed in the trichome of transgenic Nicotiana tabacum. In the next step, this construct was transformed into cotton by pollen tube pathway. PCR confirmed the integration of these two genes in the cotton genome. Cotton fiber color was observed after boll setting and it was changed from white to brown in the fiber of transgenic plants (Xu et al., 2007XU, X., WU, M., ZHAO, Q., LI, R., CHEN, J., AO, G. and YU, J., 2007. Designing and transgenic expression of melanin gene in tobacco trichome and cotton fiber. Plant Biology, vol. 9, no. 1, pp. 41-48. http://dx.doi.org/10.1055/s-2006-924346. PMid:17006798.
http://dx.doi.org/10.1055/s-2006-924346...
).
Previously used pathway is complex and involves more than one gene and steps. On the other hand, there are certain bacterial pigment production pathways that are simple and involve only one gene. One of these pathways is Rhodococcus pigment production pathway that involves pigment producing hydroxylase gene. This gene consists of 387 amino acids with 42 kDa weight. This gene converts indole into a colorless compound known as indoxyl and then converts indoxyl to indigo or indirubin (red colored) pigments. As far as gene promoter is concerned, previously used promoter LTP3 has expression window in cotton fibers from day 5 to 20. Another promoter GhSCFP has better expression window in cotton fibers from day 2 to 25. This promoter can be a better option in experiments involving expression of genes in the fibers.
In current study an attempt was made to clone and transform Rhodococcus pigment production Hydroxylase gene into C. sativa inflorescence under GhSCFP promoter and downstream Nos-terminator. The structural and functional efficiency was checked using bioinformatics tools, the cassette was synthesized from commercial source. Synthesized cassette was received pET-57 vector in lyophilized form. The cassette was then sub-cloned in plant expression vector pCAMBIA-1301. Gene construct was then transformed in C. sativa by using floral dip method. Molecular analyses were done to confirm successful integration of pigment gene into the C. sativa plants. A change in phenotype was studied for two successive generations. This is a simple and efficient transformation system which will be helpful in generation of transgenic plants with desirable traits without using antibiotic genes rather pigment production genes as a visual selection marker. This will help us to screen transgenic seeds from a large number of non-transgenic wild type seeds.
2. Materials and Methods
2.1. Designing and synthesis of pigment gene cassette
A 2581 bp cassette having ‘Rhodococcus Pigment Production Hydroxylase Gene’ (1164 bp) under a Gossypium hirsutum fiber specific promoter GhSCFP (1005bp) was designed. A Nos-terminator (252 bp) was added downstream to pigment production gene. A unique Tag (50bp) and an enhancer (58 bp) NtADH 5’ was also added in between promoter and the gene sequence (Figure 1). The resultant sequence was analyzed by using WEB-CUTTER 2.0 (2021)WEB-CUTTER 2.0. [online], 2021 [viewed 4 Aug 2021]. Available from: https://sites.unimi.it/camelot/tools/cut2.html
https://sites.unimi.it/camelot/tools/cut...
to check the cutters and non-cutter restriction enzymes.
Schematic diagram of pigment production gene cassette. The cassette consists of pigment gene regulated by GhSCFP promoter and nos3′ region.
The cassette was then subjected to bioinformatics analysis for protein, homology and phylogeny. The protein modelling was done by using SWISS-MODEL (2021a)SWISS-MODEL. [online], 2021a [viewed 4 Aug 2021]. Available from: https://swissmodel.expasy.org/interactive
https://swissmodel.expasy.org/interactiv...
tool to determine structure of the desired protein.
2.2. Protein statistics, structure and docking
DNASTAR (2021)DNASTAR. [online], 2021 [viewed 4 Aug 2021]. Available from: https://www.dnastar.com/
https://www.dnastar.com/...
was used to translate and check relative proportion of different amino acids present in the sequence. Protein secondary and tertiary structure was predicted by using SWISS-MODEL (2021b)SWISS-MODEL. [online], 2021b [viewed 4 Aug 2021]. Available from: https://swissmodel.expasy.org/
https://swissmodel.expasy.org/...
. Molecular docking of protein with substrate was checked by using Patchdock (2021a)PATCHDOCK. [online], 2021a [viewed 4 Aug 2021]. Available from: https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php
https://bioinfo3d.cs.tau.ac.il/PatchDock...
.
2.3. DNA and protein sequences and phylogenetic analysis
The DNA sequence of ‘Pigment Production Hydroxylase Gene’ (PPH_CEMB) was edited using EditSeq and SeqMan software (DNASTAR Inc., Madison, WI, USA). Initially, sequence similarity was found by BLASTn search program (NBCI, 2021NATIONAL CENTER FOR BIOTECHNOLOGY INFORMATION – NBCI. [online], 2021 [viewed 4 Aug 2021]. Available from: https://blast.ncbi.nlm.nih.gov/Blast.cgi.
https://blast.ncbi.nlm.nih.gov/Blast.cgi...
). The sequences of close relatives of PPH-CEMB were taken from GenBank (NBCI, 2021NATIONAL CENTER FOR BIOTECHNOLOGY INFORMATION – NBCI. [online], 2021 [viewed 4 Aug 2021]. Available from: https://blast.ncbi.nlm.nih.gov/Blast.cgi.
https://blast.ncbi.nlm.nih.gov/Blast.cgi...
) and included in analysis. Pairwise percentage nucleotide identity was calculated using ClustalW, available in the DNASTAR package.
Amino acid sequence of pigment production protein (PPH-CEMB) was used to blast in BLASTn search program (NBCI, 2021NATIONAL CENTER FOR BIOTECHNOLOGY INFORMATION – NBCI. [online], 2021 [viewed 4 Aug 2021]. Available from: https://blast.ncbi.nlm.nih.gov/Blast.cgi.
https://blast.ncbi.nlm.nih.gov/Blast.cgi...
) for closely related protein sequences. The amino acid sequences of close relatives of PPH_CEMB were downloaded from GenBank and included in analysis. Pairwise percentage amino acid identity was calculated using ClustalW2 (2021)CLUSTALW2. [online], 2021 [viewed 4 Aug 2021]. Available from: https://www.ebi.ac.uk/Tools/msa/clustalw2/
https://www.ebi.ac.uk/Tools/msa/clustalw...
, available in the DNASTAR package.
For phylogenetic analysis, the tree was generated using the maximum-likelihood (ML) for 1000 bootstrap iterations, at >70% support, implemented in MEGA6. The percentage of replicate trees in which the associated taxa clustered together in bootstrap test (1,000 replicates) was shown next to the nodes.
2.4. Chemical synthesis and transformation of pigment gene cassette
The designed gene cassette was synthesized from GeneScript (2021)GENESCRIPT. [online], 2021 [viewed 4 Aug 2021]. Available from: https://www.genscript.com/gene_synthesis.html
https://www.genscript.com/gene_synthesis...
in pUC57 between EcoRI and HindIII restriction sites.
Chemically competent cells of E. coli top 10 were prepared by following the already reported protocol (Chung et al., 1989CHUNG, C. T., NIEMELA, S. L. and MILLER, R. H., 1989. One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proceedings of National Academy of Sciences, USA, vol. 86, pp. 2172-2175. http://dx.doi: 10.1073/pnas.86.7.2172.
https://doi.org/http://dx.doi: 10.1073/p...
). The pUC-57 synthetic Pigment gene cassette (pUC-PPH_CEMB) was transformed into E. coli top 10 cells and plasmid was isolated by using Thermo Fisher GeneJET mini-prep kit (cat # K0503) following manufacturer’s instructions. Successful transformation of pigment gene cassette was confirmed through PCR using isolated plasmid as template.
2.5. Cloning and confirmation of pigment gene cassette (PPH_CEMB) into pCAMBIA-1301
PPH_CEMB was excised from pUC-57 using EcoR1 and HindIII. A 2581bp band of PPH_CEMB gene resolved on 1% agarose gel was excised from gel and eluted using gel elusion kit (cat # K0691). The plant expression vector pCAMBIA-1301 digested with EcoR1 and HindIII was ligated with PPH_CEMB gene between EcoR1 and HindIII sites using rapid DNA ligase kit (thermo scientific cat # K1422).
The ligation mixture was transformed into E. coli top10 competent cells and spread on LB plates (50µg/ml Tetracycline and 50µg/ml kanamycin and incubated at 37°C for overnight. Plasmid isolation was performed by using Thermo Fisher mini-prep plasmid kit (CAT #K0503). Ligation of insert was verified through restriction digestion using EcoR1 and HindIII. The successful ligation of PPH_CEMB in pCAMBIA-1301 was also confirmed through PCR amplification using plasmid DNA as a template. The internal primers 5’-ATGGACATTACTAGGACAGAGTTGA-3’ (Pigment FL Forward Primer) and 5’-TCAAAGAAGAGGGTCGGAAA-3’ (Pigment FL Reverse Primer) from the gene cassette were designed to amplify the 1164 bp region
2.6. Transformation of PPH_CEMB construct into Agrobacterium tumefaciens
The competent cells of A. tumefaciens LBA4404 were used for transformation. Electroporation and confirmation of PPH_CEMB into A. tumefaciens was performed as described previously (Puspito et al., 2015PUSPITO, A.N., RAO, A.Q., HAFEEZ, M.N., IQBAL, M.S., BAJWA, K.S., ALI, Q., RASHID, B., ABBAS, M.A., LATIF, A., SHAHID, A.A., NASIR, I.A. and HUSNAIN, T., 2015. Transformation and evaluation of Cry1Ac+ Cry2A and GTGene in Gossypium hirsutum L. Frontiers in Plant Science, vol. 6, pp. 943. http://dx.doi.org/10.3389/fpls.2015.00943. PMid:26617613.
http://dx.doi.org/10.3389/fpls.2015.0094...
).
Colony PCR was done as was performed by Iqbal et al. (2020)IQBAL, A., LATIF, A., GALBRAITH, D.W., JABBAR, B., ALI, M.A., AHMED, M., GUL, A., RAO, A.Q., SHAHID, A.A. and HUSNAIN, T., 2020. Structure-based prediction of protein–protein interactions between GhWlim5 Domain1 and GhACTIN-1 proteins: a practical evidence with improved fibre strength. Journal of Plant Biochemistry and Biotechnology, vol. 30, no. 2, pp. 373-386. http://dx.doi.org/10.1007/s13562-020-00603-7.
http://dx.doi.org/10.1007/s13562-020-006...
to confirm the transformation of PPH_CEMB construct into Agrobacterium strain LBA 4404 competent cells. The internal primers 5’-ATGGACATTACTAGGACAGAGTTGA-3’ (Pigment FL Forward Primer) and 5’-TCAAAGAAGAGGGTCGGAAA-3’ (Pigment FL Reverse Primer) from the gene cassette were designed to amplify the 1164 bp region. The PCR conditions were: initial hold at 94.0°C for 5min, second hold for 35 cycles as 94.0°C for 45sec, 55.0°C for 45sec and 72.0°C for 45sec and the final extension hold for 72.0°C for 10min. The PCR product was resolved on 1% agarose gel for 40 minutes at 100 volts and visualized under UV.
2.7. Transformation of C. sativa with PPH_CEMB using floral dip method
C. sativa seeds were obtained from Monica Schmidt’ lab, University of Arizona, USA and sown in pots filled with potting mix. Three seedlings from each pot were shifted to small pots of 11× 7.7cm in diameter and pots were kept in controlled environment till the appearance of inflorescence.
A. tumefaciens harboring PPH_CEMB gene construct was inoculated in 100 ml YEP broth having 50µg/ml kanamycin, incubated for 48 hours at 28°C in a shaking incubator. The culture was then centrifuged at 5000 rpm for 5 minutes to pellet down the cells. This pellet was then dissolved in 100 ml of 5% sucrose solution having 0.05% Silwet L-77 to break the surface tension and allow bacteria to adsorb to plant surface efficiently (Transformation solution). Flowers that started to set seeds were trimmed off from the plant by using scissors. Only emerging flowers which have higher chances to be genetically transformed were used. Transformation in C. sativa was performed by dipping flowers/inflorescence in the transformation solution for 5 minutes by applying vacuum through vacuum pump. After vacuum infiltration flowers were kept on rest for 5 minutes and opened the valve to let air get out. Opened the dome, removed plant and placed in plastic bags for overnight at dark to facilitate genetic transformation of C. sativa by Agrobacterium. Transformed plants were then shifted to growth room provided with appropriate temperature and light conditions till the onset of seeds.
2.8. Picking and sorting of seeds on the bases of color
Seeds were picked from the ripened ovaries and sorted according to color. Reddish seeds were separated from yellowish seeds. The transformed seed were packed separately in paper envelops and stored in a clean cabinet.
2.9. Sowing of seeds in pots
Reddish and yellowish seeds were sowed separately in two pots filled with peat moss. Total 20 seeds of each color (red and yellow) were sown in separate pots and kept in growth room at 25°C till the appearance of seedlings.
2.10. DNA extraction and PCR
Genomic DNA was extracted from leaves of C. sativa plants raised from yellowish and reddish seeds using the CTAB method described earlier (Doyle and Doyle, 1990DOYLE, J. J. and DOYLE, J. L., 1990. Isolation of plant DNA from fresh tissue. Focus, vol. 12, pp. 13–15.) with minor modifications. Extracted DNA was dissolved in distilled water and quality was determined by agarose gel electrophoresis, while purity with Nanodrop-ND 1000. PCR was performed using extracted DNA as a template with primers 5’-TCCAAGGGAAGATGTGAAGG-3’ (Pigment detection Forward Primer) 5’-GCCATAGCAAATGGAAGGAA-3’ (Pigment detection Reverse Primer) with the following conditions: The PCR conditions were: initial hold at 94.0°C for 5min, second hold for 35 cycles as 94.0°C for 45sec, 58.0°C for 45sec and 72.0°C for 30sec and the final extension hold for 72.0°C for 10min.
3. Results
3.1. Protein statistics
Protein statistics showed that protein consists of 387 amino acids. Out of 387 amino acids, 35 are strongly basic (+) (K, R), 49 strongly acidic (-) (D, E), 156 hydrophobic (A, I, L, F, W, V) and 82 polar amino acids (N, C, Q, S, T, Y). Amino acid and nucleotide sequence alignment is given in Figure 2. The molecular weight of pigment production hydroxylase enzyme was calculated to be 42055.71 Daltons.
Nucleotides and amino acids sequence from Rhodococcus pigment production hydroxylase gene (PPH). First line: nucleotide sequence, from 1 to 1164 is the N terminal of PPH. second line: sequence from 1 to 387 is the amino acid sequence of N terminal of pigment production gene.
3.2. Pairwise nucleotide identity
The relationship of pigment production gene (PPH_CEMB), after codon optimization to other previously reported pigment production genes was determined by subjecting, the sequence to pairwise nucleotide (nt) analysis using ClustalW, available in DNASTAR package. PPH_CEMB gene (after codon preference) showed maximum homology at 73.1% with Rhodococcus pigment gene (Accession number M55641) (Figure 3). The low nucleotide identity was expected due to change of nucleotide sequence for codon preference.
Pairwise nucleotide identity of PPH gene used in this study with Pigment genes of bacterial strains available in GenBank. PPH_CEMB gene (after codon preference) used in this study is showing maximum homology at 73.1% with Rhodococcus pigment gene (Accession number M55641). Sequences were aligned using ClustalW method in MegAlign (DNASTAR) software.
3.3. Maximum likelihood Phylogenetic Tree
Similar result was obtained for phylogenetic analysis of DNA sequence of PPH_CEMB, where PPH_CEMB was placed in distinct clade, well distant from already reported pigment gene sequences (Figure 4).
Maximum likelihood (ML) phylogenetic tree reconstructed for the pigment gene of bacterial strains available in GenBank, with representative PPH-CEMB gene (after codon preference) used in the present study. The bootstrap values are shown for nodes for 1,000 replicates.
3.4. Protein sequence and phylogenetic analysis
To confirm close association of amino acid sequence of PPH_CEMB protein with closely related amino acid sequences reported in GenBank, twenty-one close hits showing 100 percent query coverage were downloaded and used in analyses. Amino acid sequence of PPH_CEMB protein exhibited 100% amino acid homology with pigment production protein accession number P26698. The PPH_CEMB protein (used herein) clustered in same clade with pigment production protein Accession number P26698 with 99% bootstrap value (Figure 5). The results confirmed that nucleotide change for codon optimization had not changed the amino acid sequence of PPH_CEMB protein.
Maximum likelihood (ML) phylogenetic tree reconstructed for the pigment protein sequences of bacterial strains available in GenBank, with representative PPH-CEMB protein sequence used in the present study. The bootstrap values are shown for nodes of 1,000 replicates.
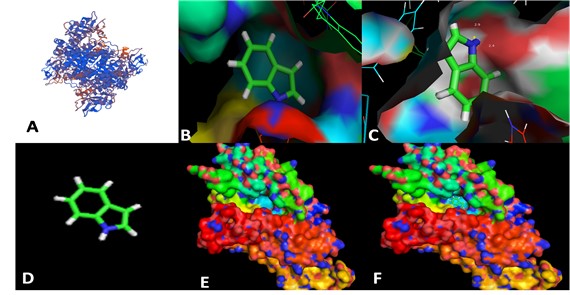
3.5. Protein structure elucidation and enzyme substrate docking
Protein SWISS-MODEL (2021a)SWISS-MODEL. [online], 2021a [viewed 4 Aug 2021]. Available from: https://swissmodel.expasy.org/interactive
https://swissmodel.expasy.org/interactiv...
predicted the possible structure of pigment production protein. The docking results indicate the global energy of -25.87, atomic contact energy (ACE) -6.52 and attractive vdW -11.23. These values suggest that the transformation of indole into pigment is favorable. Results obtained from PatchDock (2021b)PATCHDOCK. [online], 2021b [viewed 4 Aug 2021]. Available from:Available from: https://bioinfo3d.cs.tau.ac.il/PatchDock/
https://bioinfo3d.cs.tau.ac.il/PatchDock...
indicated that the indole structure fits in active site of the enzyme (Figure 6).
Structural analysis of Rhodococcus pigment production gene. A: Globular structure of pigment production hydroxylase protein (Close-up cartoon representation). B and C: the docking of indole with the pigment production protein. D: structure of indole, a substrate of pigment production enzyme. E: enzyme globular structure. F: enzyme substrate complex.
3.6. Transformation and confirmation of chemically synthesized PPH_CEMB cassette
After successfull cloning of PPH_CEMB cassette in pCAMBIA-1301, positive colonies screened by colony PCR using gene specific primers resulted in amplification of 1164 bp fragment confirmed the presence of PPH_CEMB (Figure 7).
Colony PCR for confirmation of gene cassette in pCAMBIA-1301. Lane 1 and 2: 1164bp products confirmed the successful ligation of PPH-CEMB gene cassette into pCAMBIA-1301. M: 1 kb plus DNA ladder.
3.7. Transformation into C. sativa
3.7.1. Seed sowing and shifting of seedlings in fresh pots
A patch of seedlings was observed after 4 days of sowing. Number of seedlings emerged from each pot was tabulated (Table 1). Seedlings developed inflorescence at 25 days after shifting to new pots (Table 2).
3.7.2. Transformation through Floral-Dip Method
Inflorescence was developed in 3 pots only after 25 days. These inflorescences were subjected to transformation through Floral-Dip method and left in controlled environment till the development of seeds (Figure 8, Table 3). The pots were named as PPH-1, PPH-2 and PPH-3.
Steps of Floral Dip method. A: Seeding shifted in pots. B: Inflorescence subjected to Floral-dip C: seed setting.
3.7.3. Picking and sorting of seeds
The seeds were harvested after 60 days of transformation. Total 600 seeds were separated and sorted randomly on the basis of color change. Transformation efficiency was calculated to be 1% (Table 4). The yellowish (wild type) and reddish-brown seeds were sorted (Figure 9).
Seed sorting on the basis of color change. A: wild type seeds (yellow) B: reddish brown seeds.
3.7.4. Sowing of reddish and yellowish seeds in pots and shifting of seedlings into new pots
A mat of seedlings was seen 4 days after sowing (Table 5). After 10 days of shifting of seedlings in new pots, plants developed enough foliage to be used for genomic DNA extraction (Table 6).
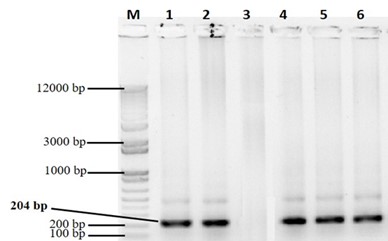
3.7.5. PCR confirmation of successful transformation of PPH_CEMB Gene
Confirmation of successful transformation of PPH_CEMB gene into C. sativa genome was done through PCR. PCR product of 204bp amplicon was obtained from red seed plants using internal detection primers 5’-ATGGACATTACTAGGACAGAGTTGA-3’ (Pigment FL Forward Primer) and 5’-TCAAAGAAGAGGGTCGGAAA-3’ (Pigment FL Reverse Primer). While no PCR amplification was seen in the plants emerged from yellow seeds (Figure 10). The plants emerged from the reddish-brown seeds were PCR positive while plant from yellow seeds were PCR negative for PPH_CEMB.
PCR confirmation of PPH-CEMB gene (204 bp) in C. sativa leaves. M: 1kb plus DNA ladder. Lane 1-2: positive control Lane 3: negative control. Lane 4-6: PCR amplification of pigment gene from RS-1, RS-2 and RS-3 lines.
4. Discussion
Genetic engineering of agronomic and model plant species has gained much importance in recent years. Usually, a gene of interest is introduced into plant in association with selectable marker gene to facilitate recovery of the transformed plants from non-transformed regenerants. Use of selectable and visual or chromogenic markers is regarded as safe and environment friendly (Pawar et al., 2010PAWAR, B., JADHAV, A., KALE, A., and CHIMOTE, V., 2010. Strategies for use of environment friendly alternative markers for detection of transformation events in transgenic plants. Asian Journal of Experimental Biological Sciences, suppl., 1-5.). Hundreds of genes have been reported to use as selectable markers or as a reporter gene for bacteria, fungi and plants such as acetolactate synthase (ALS) reviewed by (Rosellini, 2011) for development of plant herbicide tolerant, DREB2A from rice is a salt and other stress tolerant selectable marker reported by Zhu and Wu (2008)ZHU, Z. and WU, R., 2008. Regeneration of transgenic rice plants using high salt for selection without the need for antibiotics or herbicides. Plant Science, vol. 174, no. 5, pp. 519-523. http://dx.doi.org/10.1016/j.plantsci.2008.01.017.
http://dx.doi.org/10.1016/j.plantsci.200...
. In current study Rhodococcus pigment production gene was transformed in C. sativa using Agrobacterium-mediated floral dip method for the establishment of a reporter gene method of transgenic plants. The resulting changes were evaluated as reported in earlier study where this oilseed crop was successfully transformed by floral dip method (Liu et al., 2012LIU, X., BROST, J., HUTCHEON, C., GUILFOIL, R., WILSON, A.K., LEUNG, S., SHEWMAKER, C.K., ROOKE, S., NGUYEN, T., KISER, J. and DE ROCHER, J., 2012. Transformation of the oilseed crop Camelina sativa by Agrobacterium-mediated floral dip and simple large-scale screening of transformants. In Vitro Cellular & Developmental Biology - Plant, vol. 48, no. 5, pp. 462-468. http://dx.doi.org/10.1007/s11627-012-9459-7.
http://dx.doi.org/10.1007/s11627-012-945...
).
Bioinformatic analysis and codon optimization of synthetic PPH_CEMB showed 73.1% homology with Rhodococcus (M55641). The difference in nucleotide identity might be due to change of nucleotide sequence due to codon preference as reported previously (Latif et al., 2015LATIF, A., RAO, A.Q., KHAN, M.A.U., SHAHID, N., BAJWA, K.S., ASHRAF, M.A., ABBAS, M.A., AZAM, M., SHAHID, A.A., NASIR, I.A. and HUSNAIN, T., 2015. Herbicide-resistant cotton (Gossypium hirsutum) plants: an alternative way of manual weed removal. BMC Research Notes, vol. 8, pp. 453. http://dx.doi.org/10.1186/s13104-015-1397-0. PMid:26383095.
http://dx.doi.org/10.1186/s13104-015-139...
). Protein sequence analysis exhibited 100% amino acid similarity with pigment production protein (accession no P 26698). This amino acid similarity depicts that nucleotide change due to codon optimization did not affect amino acid sequence of PPH_CEMB protein. Enzyme substrate interaction using PatchDock depicts that PPH_CEMB active site is compatible to its substrate indole.
In silico cloning of synthetic PPH_CEMB cassette into pUC-57 and pCAMBIA-1301 was performed with Snap gene software (SnapGene, 2021SNAPGENE. [online], 2021 [viewed 4 Aug 2021]. Available from: https://www.snapgene.com/snapgene-viewer/
https://www.snapgene.com/snapgene-viewer...
). The chemically synthesized pUC-PPH_CEMB cassette was received in lyophilized form and plasmid construct (pUC-PPH_CEMB) harboring pigment gene was transformed into E. coli Top10 competent cells and presence of pigment gene was confirmed through PCR. The PPH_CEMB cassette was cloned into pCAMBIA-1301 and cloning was confirmed through colony PCR and restriction digestion using EcoRI and HindIII. The resulting plasmid pCAMBIA-1301_PPH_CEMB was transformed into A. tumefaciens LBA4404 competent cells through electroporation. Successful electroporation was confirmed through colony PCR.
Total 19 out of 600 hundred seeds showed a change in color from yellow to reddish brown. These results coincide with earlier studies in which transformation of β-carotene gene in C. sativa using floral dip method changed seed color from yellow to orange (Schmidt and Pendarvis, 2017SCHMIDT, M.A. and PENDARVIS, K., 2017. Proteome rebalancing in transgenic Camelina occurs within the enlarged proteome induced by β-carotene accumulation and storage protein suppression. Transgenic Research, vol. 26, no. 2, pp. 171-186. http://dx.doi.org/10.1007/s11248-016-9992-y. PMid:27771868.
http://dx.doi.org/10.1007/s11248-016-999...
). The overall transformation efficiency calculated on the basis of color change was 1%, that is close (0.8%) to already reported transformation in C. sativa and cotton (Liu et al., 2012LIU, X., BROST, J., HUTCHEON, C., GUILFOIL, R., WILSON, A.K., LEUNG, S., SHEWMAKER, C.K., ROOKE, S., NGUYEN, T., KISER, J. and DE ROCHER, J., 2012. Transformation of the oilseed crop Camelina sativa by Agrobacterium-mediated floral dip and simple large-scale screening of transformants. In Vitro Cellular & Developmental Biology - Plant, vol. 48, no. 5, pp. 462-468. http://dx.doi.org/10.1007/s11627-012-9459-7.
http://dx.doi.org/10.1007/s11627-012-945...
; Ahmed et al., 2020AHMED, M., IQBAL, A., LATIF, A., UD DIN, S., SARWAR, M.B., WANG, X., RAO, A.Q., HUSNAIN, T. and SHAHID, A.A., 2020. Overexpression of a sucrose synthase gene indirectly improves cotton fiber quality through sucrose cleavage. Frontiers in Plant Science, vol. 11, pp. 476251. http://dx.doi.org/10.3389/fpls.2020.476251. PMid:33281834.
http://dx.doi.org/10.3389/fpls.2020.4762...
; Iqbal et al., 2020IQBAL, A., LATIF, A., GALBRAITH, D.W., JABBAR, B., ALI, M.A., AHMED, M., GUL, A., RAO, A.Q., SHAHID, A.A. and HUSNAIN, T., 2020. Structure-based prediction of protein–protein interactions between GhWlim5 Domain1 and GhACTIN-1 proteins: a practical evidence with improved fibre strength. Journal of Plant Biochemistry and Biotechnology, vol. 30, no. 2, pp. 373-386. http://dx.doi.org/10.1007/s13562-020-00603-7.
http://dx.doi.org/10.1007/s13562-020-006...
). Selection of transgenic C. sativa on the basis of color change was used to differentiate between transgenic and non-transgenic plants. Same strategy was used by Schmidt and Pendarvis (2017)SCHMIDT, M.A. and PENDARVIS, K., 2017. Proteome rebalancing in transgenic Camelina occurs within the enlarged proteome induced by β-carotene accumulation and storage protein suppression. Transgenic Research, vol. 26, no. 2, pp. 171-186. http://dx.doi.org/10.1007/s11248-016-9992-y. PMid:27771868.
http://dx.doi.org/10.1007/s11248-016-999...
in selection of transgenic C. sativa. The Rhodococcus pigment production Hyrdoylase gene is responsible for the development of indigo/indirubin pigments in bacteria. This small pigment gene may be helpful for construction of a new generation of chromogenic vectors which do not need costly substrates like X-Gluc (5-Bromo-4-chloro-1H-indol-3-yl β-D-glucopyranosiduronic acid) for detection of gene expression. Rhodococcus pigment gene can be used as visual marker in place of selectable marker gene (antibiotic resistance gene) to eliminate antibiotic selection. The change in seed color is safe to screen putative transgenic plants as compared to use of antibiotic as selection drug. Antibiotic selection markers often remain incorporated into the genome of transgenic plants and often poses bio-safety concern by its probability of spreading antibiotic resistance among microbes through horizontal gene transfer (HGT) (Keese, 2008KEESE, P., 2008. Risks from GMOs due to horizontal gene transfer. Environmental Biosafety Research, vol. 7, no. 3, pp. 123-149. http://dx.doi.org/10.1051/ebr:2008014. PMid:18801324.
http://dx.doi.org/10.1051/ebr:2008014...
). Similar work on reporter gene of plant selectable markers have been reported by a number of scientists around the world such as Enolpyruvylshikimate-3-phosphate synthase-EPSPS, Charng et al. (2008)CHARNG, Y., LI, K., TAI, H., LIN and TU, 2008. An inducible transposon system to terminate the function of a selectable marker in transgenic plants. Molecular Breeding, vol. 213, pp. 59–368. http://dx. DOI 10.1007/s11032-007-9137-3.
https://doi.org/http://dx. DOI 10.1007/s...
, D-serine ammonia lyase-dsdA Erikson et al. (2005)ERIKSON, O., HERTZBERG, M. and NASHOLM, T., 2005. The dsdA gene from Escherichia coli provides a novel selectable marker for plant transformation. Plant Molecular Biology, vol. 57, no. 3, pp. 425-433. http://dx.doi.org/10.1007/s11103-004-7902-9. PMid:15830131.
http://dx.doi.org/10.1007/s11103-004-790...
, glutathione S-transferase-GST Milligan et al. (2001)MILLIGAN, A.S., DALY, A., PARRY, M.A.J., LAZZERI, P.A. and JEPSON, I., 2001. The expression of a maize glutathione S-transferase gene in transgenic wheat confers herbicide tolerance, both in planta and in vitro. Molecular Breeding, vol. 7, no. 4, pp. 301-315. http://dx.doi.org/10.1023/A:1011652821765.
http://dx.doi.org/10.1023/A:101165282176...
and Heat shock protein101-HSP101 Chang et al. (2007)CHANG, C.C., HUANG, P.S., LIN, H.R. and LU, C.H., 2007. Transactivation of protein expression by rice HSP101 in planta and using Hsp101 as a selection marker for transformation. Plant & Cell Physiology, vol. 48, no. 8, pp. 1098-1107. http://dx.doi.org/10.1093/pcp/pcm080. PMid:17597080.
http://dx.doi.org/10.1093/pcp/pcm080...
. Plant scientists need more and more such reporter and environment friendly markers genes and the current study is one such effort.
5. Future Prospects of the Research
Cotton fiber quality is a multigenic trait. The PPH_CEMB construct, in combination of some other genes, will be transformed into cotton plants for the development of colored cotton fiber.
Acknowledgements
We extent our gratitude to the Dr. Monica Schmidt Lab, School of Plant Sciences, University of Arizona, USA for providing the seeds of Camelina sativa. Authors are also thankful to Higher Education Commission (HEC) of Pakistan for providing financial support
References
- AHMED, M., IQBAL, A., LATIF, A., UD DIN, S., SARWAR, M.B., WANG, X., RAO, A.Q., HUSNAIN, T. and SHAHID, A.A., 2020. Overexpression of a sucrose synthase gene indirectly improves cotton fiber quality through sucrose cleavage. Frontiers in Plant Science, vol. 11, pp. 476251. http://dx.doi.org/10.3389/fpls.2020.476251 PMid:33281834.
» http://dx.doi.org/10.3389/fpls.2020.476251 - BEYER, P., AL-BABILI, S., YE, X., LUCCA, P., SCHAUB, P., WELSCH, R. and POTRYKUS, I., 2002. Golden rice: introducing the β-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency. The Journal of Nutrition, vol. 132, no. 3, pp. 506S-510S. http://dx.doi.org/10.1093/jn/132.3.506S PMid:11880581.
» http://dx.doi.org/10.1093/jn/132.3.506S - CHANG, C.C., HUANG, P.S., LIN, H.R. and LU, C.H., 2007. Transactivation of protein expression by rice HSP101 in planta and using Hsp101 as a selection marker for transformation. Plant & Cell Physiology, vol. 48, no. 8, pp. 1098-1107. http://dx.doi.org/10.1093/pcp/pcm080 PMid:17597080.
» http://dx.doi.org/10.1093/pcp/pcm080 - CHARNG, Y., LI, K., TAI, H., LIN and TU, 2008. An inducible transposon system to terminate the function of a selectable marker in transgenic plants. Molecular Breeding, vol. 213, pp. 59–368. http://dx. DOI 10.1007/s11032-007-9137-3.
» https://doi.org/http://dx. DOI 10.1007/s11032-007-9137-3 - CLUSTALW2. [online], 2021 [viewed 4 Aug 2021]. Available from: https://www.ebi.ac.uk/Tools/msa/clustalw2/
» https://www.ebi.ac.uk/Tools/msa/clustalw2/ - CHUNG, C. T., NIEMELA, S. L. and MILLER, R. H., 1989. One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proceedings of National Academy of Sciences, USA, vol. 86, pp. 2172-2175. http://dx.doi: 10.1073/pnas.86.7.2172.
» https://doi.org/http://dx.doi: 10.1073/pnas.86.7.2172 - COSTA-FONT, M., GIL, J.M. and TRAILL, W.B., 2008. Consumer acceptance, valuation of and attitudes towards genetically modified food: review and implications for food policy. Food Policy, vol. 33, no. 2, pp. 99-111. http://dx.doi.org/10.1016/j.foodpol.2007.07.002
» http://dx.doi.org/10.1016/j.foodpol.2007.07.002 - DNASTAR. [online], 2021 [viewed 4 Aug 2021]. Available from: https://www.dnastar.com/
» https://www.dnastar.com/ - DOYLE, J. J. and DOYLE, J. L., 1990. Isolation of plant DNA from fresh tissue. Focus, vol. 12, pp. 13–15.
- ERIKSON, O., HERTZBERG, M. and NASHOLM, T., 2005. The dsdA gene from Escherichia coli provides a novel selectable marker for plant transformation. Plant Molecular Biology, vol. 57, no. 3, pp. 425-433. http://dx.doi.org/10.1007/s11103-004-7902-9 PMid:15830131.
» http://dx.doi.org/10.1007/s11103-004-7902-9 - GENESCRIPT. [online], 2021 [viewed 4 Aug 2021]. Available from: https://www.genscript.com/gene_synthesis.html
» https://www.genscript.com/gene_synthesis.html - GOULD, J.H. and MAGALLANES-CEDENO, M., 1998. Adaptation of cotton shoot apex culture to Agrobacterium-mediated transformation. Plant Molecular Biology Reporter, vol. 16, no. 3, pp. 283-283. http://dx.doi.org/10.1023/A:1007438104369
» http://dx.doi.org/10.1023/A:1007438104369 - IQBAL, A., LATIF, A., GALBRAITH, D.W., JABBAR, B., ALI, M.A., AHMED, M., GUL, A., RAO, A.Q., SHAHID, A.A. and HUSNAIN, T., 2020. Structure-based prediction of protein–protein interactions between GhWlim5 Domain1 and GhACTIN-1 proteins: a practical evidence with improved fibre strength. Journal of Plant Biochemistry and Biotechnology, vol. 30, no. 2, pp. 373-386. http://dx.doi.org/10.1007/s13562-020-00603-7
» http://dx.doi.org/10.1007/s13562-020-00603-7 - KEESE, P., 2008. Risks from GMOs due to horizontal gene transfer. Environmental Biosafety Research, vol. 7, no. 3, pp. 123-149. http://dx.doi.org/10.1051/ebr:2008014 PMid:18801324.
» http://dx.doi.org/10.1051/ebr:2008014 - LATIF, A., RAO, A.Q., KHAN, M.A.U., SHAHID, N., BAJWA, K.S., ASHRAF, M.A., ABBAS, M.A., AZAM, M., SHAHID, A.A., NASIR, I.A. and HUSNAIN, T., 2015. Herbicide-resistant cotton (Gossypium hirsutum) plants: an alternative way of manual weed removal. BMC Research Notes, vol. 8, pp. 453. http://dx.doi.org/10.1186/s13104-015-1397-0 PMid:26383095.
» http://dx.doi.org/10.1186/s13104-015-1397-0 - LIU, X., BROST, J., HUTCHEON, C., GUILFOIL, R., WILSON, A.K., LEUNG, S., SHEWMAKER, C.K., ROOKE, S., NGUYEN, T., KISER, J. and DE ROCHER, J., 2012. Transformation of the oilseed crop Camelina sativa by Agrobacterium-mediated floral dip and simple large-scale screening of transformants. In Vitro Cellular & Developmental Biology - Plant, vol. 48, no. 5, pp. 462-468. http://dx.doi.org/10.1007/s11627-012-9459-7
» http://dx.doi.org/10.1007/s11627-012-9459-7 - MILLIGAN, A.S., DALY, A., PARRY, M.A.J., LAZZERI, P.A. and JEPSON, I., 2001. The expression of a maize glutathione S-transferase gene in transgenic wheat confers herbicide tolerance, both in planta and in vitro. Molecular Breeding, vol. 7, no. 4, pp. 301-315. http://dx.doi.org/10.1023/A:1011652821765
» http://dx.doi.org/10.1023/A:1011652821765 - NATIONAL CENTER FOR BIOTECHNOLOGY INFORMATION – NBCI. [online], 2021 [viewed 4 Aug 2021]. Available from: https://blast.ncbi.nlm.nih.gov/Blast.cgi
» https://blast.ncbi.nlm.nih.gov/Blast.cgi - NIELSEN, K.M., BONES, A.M., SMALLA, K. and VAN ELSAS, J.D., 1998. Horizontal gene transfer from transgenic plants to terrestrial bacteria–a rare event? FEMS Microbiology Reviews, vol. 22, no. 2, pp. 79-103. http://dx.doi.org/10.1016/S0168-6445(98)00009-6 PMid:9729765.
» http://dx.doi.org/10.1016/S0168-6445(98)00009-6 - PATCHDOCK. [online], 2021a [viewed 4 Aug 2021]. Available from: https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php
» https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php - PATCHDOCK. [online], 2021b [viewed 4 Aug 2021]. Available from:Available from: https://bioinfo3d.cs.tau.ac.il/PatchDock/
» https://bioinfo3d.cs.tau.ac.il/PatchDock/ - PAWAR, B., JADHAV, A., KALE, A., and CHIMOTE, V., 2010. Strategies for use of environment friendly alternative markers for detection of transformation events in transgenic plants. Asian Journal of Experimental Biological Sciences, suppl., 1-5.
- PONTIROLI, A., SIMONET, P., FROSTEGARD, A., VOGEL, T.M. and MONIER, J.-M., 2007. Fate of transgenic plant DNA in the environment. Environmental Biosafety Research, vol. 6, no. 1-2, pp. 15-35. http://dx.doi.org/10.1051/ebr:2007037 PMid:17961478.
» http://dx.doi.org/10.1051/ebr:2007037 - PUSPITO, A.N., RAO, A.Q., HAFEEZ, M.N., IQBAL, M.S., BAJWA, K.S., ALI, Q., RASHID, B., ABBAS, M.A., LATIF, A., SHAHID, A.A., NASIR, I.A. and HUSNAIN, T., 2015. Transformation and evaluation of Cry1Ac+ Cry2A and GTGene in Gossypium hirsutum L. Frontiers in Plant Science, vol. 6, pp. 943. http://dx.doi.org/10.3389/fpls.2015.00943 PMid:26617613.
» http://dx.doi.org/10.3389/fpls.2015.00943 - RAO, A.Q., IRFAN, M., SALEEM, Z., NASIR, I.A., RIAZUDDIN, S. and HUSNAIN, T., 2011. Overexpression of the phytochrome B gene from Arabidopsis thaliana increases plant growth and yield of cotton (Gossypium hirsutum). Journal of Zhejiang University. Science. B., vol. 12, no. 4, pp. 326-334. http://dx.doi.org/10.1631/jzus.B1000168 PMid:21462389.
» http://dx.doi.org/10.1631/jzus.B1000168 - ROSELLINI, D., 2012. Selectable Markers and Reporter Genes: A Well-Furnished Toolbox for Plant Science and Genetic Engineering. Critical Reviews in Plant Sciences, vol. 31, no. 5, pp. 401-453. https://doi.org/10.1080/07352689.2012.683373
» https://doi.org/10.1080/07352689.2012.683373 - SCHMIDT, M.A. and PENDARVIS, K., 2017. Proteome rebalancing in transgenic Camelina occurs within the enlarged proteome induced by β-carotene accumulation and storage protein suppression. Transgenic Research, vol. 26, no. 2, pp. 171-186. http://dx.doi.org/10.1007/s11248-016-9992-y PMid:27771868.
» http://dx.doi.org/10.1007/s11248-016-9992-y - SNAPGENE. [online], 2021 [viewed 4 Aug 2021]. Available from: https://www.snapgene.com/snapgene-viewer/
» https://www.snapgene.com/snapgene-viewer/ - SWISS-MODEL. [online], 2021a [viewed 4 Aug 2021]. Available from: https://swissmodel.expasy.org/interactive
» https://swissmodel.expasy.org/interactive - SWISS-MODEL. [online], 2021b [viewed 4 Aug 2021]. Available from: https://swissmodel.expasy.org/
» https://swissmodel.expasy.org/ - TRAAVIK, T., and CHING, L.L., 2009. Biosafety first: holistic approaches to risk and uncertainty in genetic engineering and genetically modified organisms. Malaysia: Third World Network and GenØk.
- VAN DEN EEDE, G., AARTS, H., BUHK, H.-J., CORTHIER, G., FLINT, H.J., HAMMES, W., JACOBSEN, B., MIDTVEDT, T., VAN DER VOSSEN, J., VON WRIGHT, A., WACKERNAGEL, W. and WILCKS, A., 2004. The relevance of gene transfer to the safety of food and feed derived from genetically modified (GM) plants. Food and Chemical Toxicology, vol. 42, no. 7, pp. 1127-1156. http://dx.doi.org/10.1016/j.fct.2004.02.001 PMid:15123384.
» http://dx.doi.org/10.1016/j.fct.2004.02.001 - WEB-CUTTER 2.0. [online], 2021 [viewed 4 Aug 2021]. Available from: https://sites.unimi.it/camelot/tools/cut2.html
» https://sites.unimi.it/camelot/tools/cut2.html - XU, X., WU, M., ZHAO, Q., LI, R., CHEN, J., AO, G. and YU, J., 2007. Designing and transgenic expression of melanin gene in tobacco trichome and cotton fiber. Plant Biology, vol. 9, no. 1, pp. 41-48. http://dx.doi.org/10.1055/s-2006-924346 PMid:17006798.
» http://dx.doi.org/10.1055/s-2006-924346 - ZHU, Z. and WU, R., 2008. Regeneration of transgenic rice plants using high salt for selection without the need for antibiotics or herbicides. Plant Science, vol. 174, no. 5, pp. 519-523. http://dx.doi.org/10.1016/j.plantsci.2008.01.017
» http://dx.doi.org/10.1016/j.plantsci.2008.01.017
Publication Dates
-
Publication in this collection
27 Apr 2022 -
Date of issue
2024
History
-
Received
04 Aug 2021 -
Accepted
15 Mar 2022