Abstract
Ligula intestinalis is a cestode parasite that affects freshwater fish in different countries of the world. The current study aims to reveal the phylogenetic, genetic and haplotype diversity of mt-CO1 gene sequences sent to the NCBI database from different countries by using in-silico analysis. The 105 mt-CO1 (371 bp) gene sequences of L. intestinalis obtained from NCBI were used for bioinformatics analyses. Sequences were subjected to phylogenetic and haplotype analysis. As a result of the haplotype analysis of L. intestinalis, 38 haplotypes were obtained from 13 different countries. Hap24 constituted 44.76% of the obtained haplotype network. Changes in nucleotides between haplotypes occurred at 1-84 different points. China and Turkey have highest fixation index (Fst) values of 0.59761, while the lowest (-0.10526) was found between Russia and Turkey. This study provides a baseline for future studies on extensive scale on the epidemiology, ecological aspects, distribution pattern, transmission dynamics and population dispersion of L. intestinalis worldwide.
Keywords:
Ligula intestinalis; genetic variation; mt-CO1; in-silico analysis
Resumo
Ligula intestinalis é um parasita cestódeo que acomete peixes de água doce em diversos países do mundo. O presente estudo visa revelar a diversidade filogenética, genética e de haplótipos das sequências do gene mt-CO1 enviadas ao banco de dados do NCBI de diferentes países, por meio de análise in-silico. As sequências gênicas de 105 mt-CO1 (371 pb) de L. intestinalis obtidas do NCBI foram utilizadas para análises bioinformáticas. As sequências foram submetidas a análise filogenética e de haplótipos. Como resultado da análise de haplótipos de L. intestinalis, 38 haplótipos foram obtidos de 13 países diferentes. Hap24 constituiu 44,76% da rede de haplótipos obtida. Mudanças nos nucleotídeos entre os haplótipos ocorreram em 1-84 pontos diferentes. A China e a Turquia apresentam os maiores valores do índice de fixação (Fst), 0,59761, enquanto o menor (-0,10526) foi encontrado entre a Rússia e a Turquia. Este estudo fornece uma linha de base para futuros estudos em larga escala sobre epidemiologia, aspectos ecológicos, padrão de distribuição, dinâmica de transmissão e dispersão populacional de L. intestinalis em todo o mundo.
Palavras-chave:
Ligula intestinalis; variação genética; mt-CO1; análise in-silico
1. Introduction
Ligula intestinalis is a pseudophyllidean cestode, which is located in the Diphyllobothridae family. It is a common parasitic disease that causes the most important health problems in fish farming. The final hosts of L. intestinalis are waterfowls while the first intermediate host is Crustacea, and the second intermediate host is freshwater fish (Hoole et al., 2010HOOLE, D., CARTER, V. and DUFOUR, S., 2010. Ligula intestinalis (cestoda: Pseudophyllidea): an ideal fish-metazoan parasite model? Parasitology, vol. 137, no. 3, pp. 425-438. http://dx.doi.org/10.1017/S0031182010000107. PMid:20163752.
http://dx.doi.org/10.1017/S0031182010000...
). The eggs, which are excreted in feces of final hosts, develop and hatch in water as coracidium. Free coracidiums entered in first intermediate host from drinking of contaminated water, the crustacean copepods, and develop into procercoids in the abdominal cavities within 2-3 weeks. The second intermediate host ingests copepods containing procercoids. Then procercoids transform into the plerocercoids in the abdominal cavity of the fish. In the waterfowl that eat fish contaminated with plerocercoid, the larvae mature into the adult parasite (Loot et al., 2002LOOT, G., AULAGNIER, S., LEK, S., THOMAS, F. and GUÉGAN, J.F., 2002. Experimental demonstration of a behavioural modification in a cyprinid fish, Rutilus rutilus (L.), induced by a parasite, Ligula intestinalis (L.). Canadian Journal of Zoology, vol. 80, no. 4, pp. 738-744. http://dx.doi.org/10.1139/z02-043.
http://dx.doi.org/10.1139/z02-043...
).
The most prominent parasitism phase of L. intestinalis during its life cycle is the plerocercoid stage. L. intestinalis causes important effects such as physical damage, growth retardation, changes in blood parameters, immunological disorders, hormonal disruption and behavioral changes during the development in the abdominal cavity of the fish (Loot et al., 2001LOOT, G., FRANCISCO, P., SANTOUL, F., LEK, S. and GUÉGAN, J.F., 2001. The three hosts of the Ligula intestinalis (Cestoda) life cycle in Lavernose-Lacasse gravel pit, France. Archiv für Hydrobiologie, vol. 152, no. 3, pp. 511-525. http://dx.doi.org/10.1127/archiv-hydrobiol/152/2001/511.
http://dx.doi.org/10.1127/archiv-hydrobi...
; Barber et al., 2000BARBER, I., HOARE, D. and KRAUSE, J., 2000. Effects of parasites on fish behaviour: a review and evolutionary perspective. Reviews in Fish Biology and Fisheries, vol. 10, no. 2, pp. 131-165. http://dx.doi.org/10.1023/A:1016658224470.
http://dx.doi.org/10.1023/A:101665822447...
; Britton et al., 2009BRITTON, J.R., JACKSON, M.C. and HARPER, D.M., 2009. Ligula intestinalis (Cestoda: Diphyllobothriidae) in Kenya: a field investigation into host specificity and behavioural alterations. Parasitology, vol. 136, no. 11, pp. 1367-1373. http://dx.doi.org/10.1017/S003118200999059X. PMid:19627635.
http://dx.doi.org/10.1017/S0031182009990...
). The prevalence of L. intestinalis in fish species changes according to the regions. The prevalance was 7.2% in Zimbabwe (Barson and Marshall, 2003BARSON, M. and MARSHALL, B., 2003. The occurrence of the tapeworm Ligula intestinalis (L.), in Barbus paludinosus from a small dam in Zimbabwe. African Journal of Aquatic Science, vol. 28, no. 2, pp. 175-178. http://dx.doi.org/10.2989/16085910309503782.
http://dx.doi.org/10.2989/16085910309503...
), 29.00% in Ethiopia (Emaminew et al., 2014EMAMINEW, T., DEREJE, B. and ABDU, M., 2014. Prevalence of Ligula intestinalis larvae in Barbus fish genera at Lake Tana, Ethiopia. World Journal of Fish and Marine Sciences, vol. 6, no. 5, pp. 408-416.), 14.98% in Malawi (Gabagambi and Skorping, 2018GABAGAMBI, N.P. and SKORPING, A., 2018. Spatial and temporal distribution of Ligula intestinalis (Cestoda: Diphyllobothriidea) in usipa (Engraulicypris sardella)(Pisces: Cyprinidae) in Lake Nyasa. Journal of Helminthology, vol. 92, no. 4, pp. 410-416. http://dx.doi.org/10.1017/S0022149X17000724. PMid:28818117.
http://dx.doi.org/10.1017/S0022149X17000...
), 9.7% in Iran (Shargh et al., 2008SHARGH, S., SHAMSAII, M. and KARIMI, S., 2008. Distribution of parasitic Cestod “Ligula intestinalis” in Mazandaran region. Iranian Journal of Parasitology, vol. 3, no. 2, pp. 26-33.), 23.38% in Sri Lanka (Weliange and Amarasinghe, 2001WELIANGE, W. and AMARASINGHE, U., 2001. The occurrence of cestode Ligula intestinalis (Linnaeus) from attentive carplet Amblypharyngodon melettinus (Valenciennes) in Sri Lanka. Asian Fisheries Science, vol. 14, no. 1, pp. 95-98. http://dx.doi.org/10.33997/j.afs.2001.14.1.011.
http://dx.doi.org/10.33997/j.afs.2001.14...
). The prevalence rate of L. intestinalis in fish in Turkey was 23% in Balikesir (Koc et al., 2006KOC, H.T., ERDOGAN, Z. and COZ-RAKOVAC, R., 2006. The occurrence of Ligula intestinalis (L.) observed in chub (Leuciscus cephalus L.) from Caparlipatlak Dam lake, Ivrindi-Balikesir, Turkey. Periodicum Biologorum, vol. 108, no. 2, pp. 183-187.), 71.5% in Eskisehir (Ozbek and Ozturk, 2010OZBEK, M. and OZTURK, M.O., 2010. Investigations on Ligula intestinalis plerocercoid L., 1758 infection of some fishes from Dam lake Kunduzlar (Kirka, Eskişehir). Turkiye Parazitoloji Dergisi, vol. 34, no. 2, pp. 112-117. PMid:20597057.) and 73.33% in Tokat provinces (Turgut et al., 2011TURGUT, E., DEVELI, N., YESILAYER, N. and BUHAN, E., 2011. Seasonal occurrence of Ligula intestinalis infection in cyprinids from Almus Dam Lake (Turkey). KSÜ Doğa Bilimleri Dergisi, vol. 14, no. 3, pp. 9-11.).
Ligulosis is a foodborne zoonotic disease. There are various reports regarding this situation (Urdes and Hangan, 2013URDES, L.D. and HANGAN, M., 2013. The epidemiology of Ligula intestinalis (Phylum Platyhelminthes) within the cyprinid populations inhabiting the Danubian Delta area. Scientific Papers Animal Science and Biotechnologies, vol. 46, no. 1, pp. 273-276.). Fish production has a special place in trade and economy for the human population, more importantly, there is a greater need in tropical and subtropical countries with nutritional deficiency problems. Parasites that cause diseases in fish cause great economic losses as mortality increases, while expenses incurred for treatment increase farm costs, and a decrease in growth rate depending on the periods of parasitic diseases causes weight loss accordingly. All these situations hinder the progress of fish farms (Osuigwe and Obiekezie, 2007OSUIGWE, D. and OBIEKEZIE, A., 2007. Assessment of the growth performance and feed utilization of fingering Heterobranchus longifilis fed raw and boiled jackbean (Canavalia ensiformis) seed meal as fishmeal substitute. Journal of Fisheries International, vol. 2, pp. 37-41.).
Due to the advancement of today's technology and advances in sequencing and computational technologies in the genetic field, the obtained DNA sequences have become a source of information to advance the understanding between genetic relationships and to clarify the evolutionary process (Tibayrenc, 2005TIBAYRENC, M., 2005. Bridging the gap between molecular epidemiologists and evolutionists. Trends in Microbiology, vol. 13, no. 12, pp. 575-580. http://dx.doi.org/10.1016/j.tim.2005.09.004. PMid:16214342.
http://dx.doi.org/10.1016/j.tim.2005.09....
).
It has come ready to contribute to taxonomic research, population genetics and phylogenetic by developing DNA barcodes, which are short DNA sequences of a standard gene region. At the same time, many groups around the world are working day by day to obtain an equipped DNA barcode database that covers most of the world's biodiversity (Ali et al., 2014ALI, M.A., GYULAI, G., HIDVÉGI, N., KERTI, B., HEMAID, F.M., PANDEY, A.K. and LEE, J., 2014. The changing epitome of species identification–DNA barcoding. Saudi Journal of Biological Sciences, vol. 21, no. 3, pp. 204-231. http://dx.doi.org/10.1016/j.sjbs.2014.03.003. PMid:24955007.
http://dx.doi.org/10.1016/j.sjbs.2014.03...
; Hajibabaei et al., 2007HAJIBABAEI, M., SINGER, G.A., HEBERT, P.D. and HICKEY, D.A., 2007. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends in Genetics, vol. 23, no. 4, pp. 167-172. http://dx.doi.org/10.1016/j.tig.2007.02.001. PMid:17316886.
http://dx.doi.org/10.1016/j.tig.2007.02....
; Sarkar and Trizna, 2011SARKAR, I.N. and TRIZNA, M., 2011. The Barcode of Life Data Portal: bridging the biodiversity informatics divide for DNA barcoding. PLoS One, vol. 6, no. 7, p. e14689. http://dx.doi.org/10.1371/journal.pone.0014689. PMid:21818249.
http://dx.doi.org/10.1371/journal.pone.0...
). It enables species identification and classification by obtaining nucleotide sequences of the mitochondrial Cytochrome Oxidase subunit 1 (CO1) region of the genome for DNA barcoding purposes (Hebert et al., 2003HEBERT, P.D., CYWINSKA, A., BALL, S.L. and DEWAARD, J.R., 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B, vol. 270, no. 1512, pp. 313-321. http://dx.doi.org/10.1098/rspb.2002.2218. PMid:12614582.
http://dx.doi.org/10.1098/rspb.2002.2218...
). Present study was aimed to investigate the haplotype diversity, genetic variation, and phylogeny of L. intestinalis by using mt-CO1 genetic marker deposited to GenBank from different geographical zones.
2. Methodology
2.1. Collection of data
The sequences of the partial mt-CO1 gene fragment of L. intestinalis had been submitted to the National Center for Biotechnology Information, USA, (NCBI) (www.ncbi.nlm.nih.gov) until May 05, 2021 were used for the bioinformatics analyses. A total of 105 sequences were identified after searching these gene sequences in the NCBI database. The sequences of L. intestinalis were used from different countries of Europe, Asia, Africa and North America while the selected sequences were retrieved by focusing on gene region, host and location.
2.2. Data and phylogenetic analysis
The selected sequences were retrieved by using FASTA format from NCBI database to the CLC Sequence Viewer 8 (QIAGEN CLC Main Workbench, 2007QIAGEN CLC MAIN WORKBENCH, 2007. QIAGEN CLC Main Workbench: the user-friendly solution for basic sequencing analysis [software]. Hilden: QIAGEN.). A reference sequence (Access no. NC039445) of L. intestinalis was used to align all obtained sequences of different lengths. After all sequences were trimmed and aligned, 105 sequences with a length of 371 bp were obtained and then used for bioinformatics analyses. Following, the phylogenetic tree was created with the Neighbour Joining (NJ) model. For statistical support to specific branches, 1000 bootstrap replicates were obtained.
2.3. Haplotype analysis
After all sequences were saved in FASTA format then DnaSP 6 program was used for the analyses. With the program, statistics about haplotype and nucleotide change values, haplotype numbers, nucleotide content, and mutation amounts between haplotypes were determined (Rozas et al., 2017ROZAS, J., FERRER-MATA, A., SÁNCHEZ-DELBARRIO, J.C., GUIRAO-RICO, S., LIBRADO, P., RAMOS-ONSINS, S.E. and SÁNCHEZ-GRACIA, A., 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, vol. 34, no. 12, pp. 3299-3302. http://dx.doi.org/10.1093/molbev/msx248. PMid:29029172.
http://dx.doi.org/10.1093/molbev/msx248...
). In the DnaSP6 tool, it was also recorded in NEXUS format to create the haplotype network with sequences (Maddison et al., 1997MADDISON, D.R., SWOFFORD, D.L. and MADDISON, W.P., 1997. NEXUS: an extensible file format for systematic information. Systematic Biology, vol. 46, no. 4, pp. 590-621. http://dx.doi.org/10.1093/sysbio/46.4.590. PMid:11975335.
http://dx.doi.org/10.1093/sysbio/46.4.59...
). Then, the haplotype network was created by using with PopART (Population Analysis with Reticulate Trees) program (Leigh and Bryant, 2015LEIGH, J.W. and BRYANT, D., 2015. Popart: full‐feature software for haplotype network construction. Methods in Ecology and Evolution, vol. 6, no. 9, pp. 1110-1116. http://dx.doi.org/10.1111/2041-210X.12410.
http://dx.doi.org/10.1111/2041-210X.1241...
).
3. Results
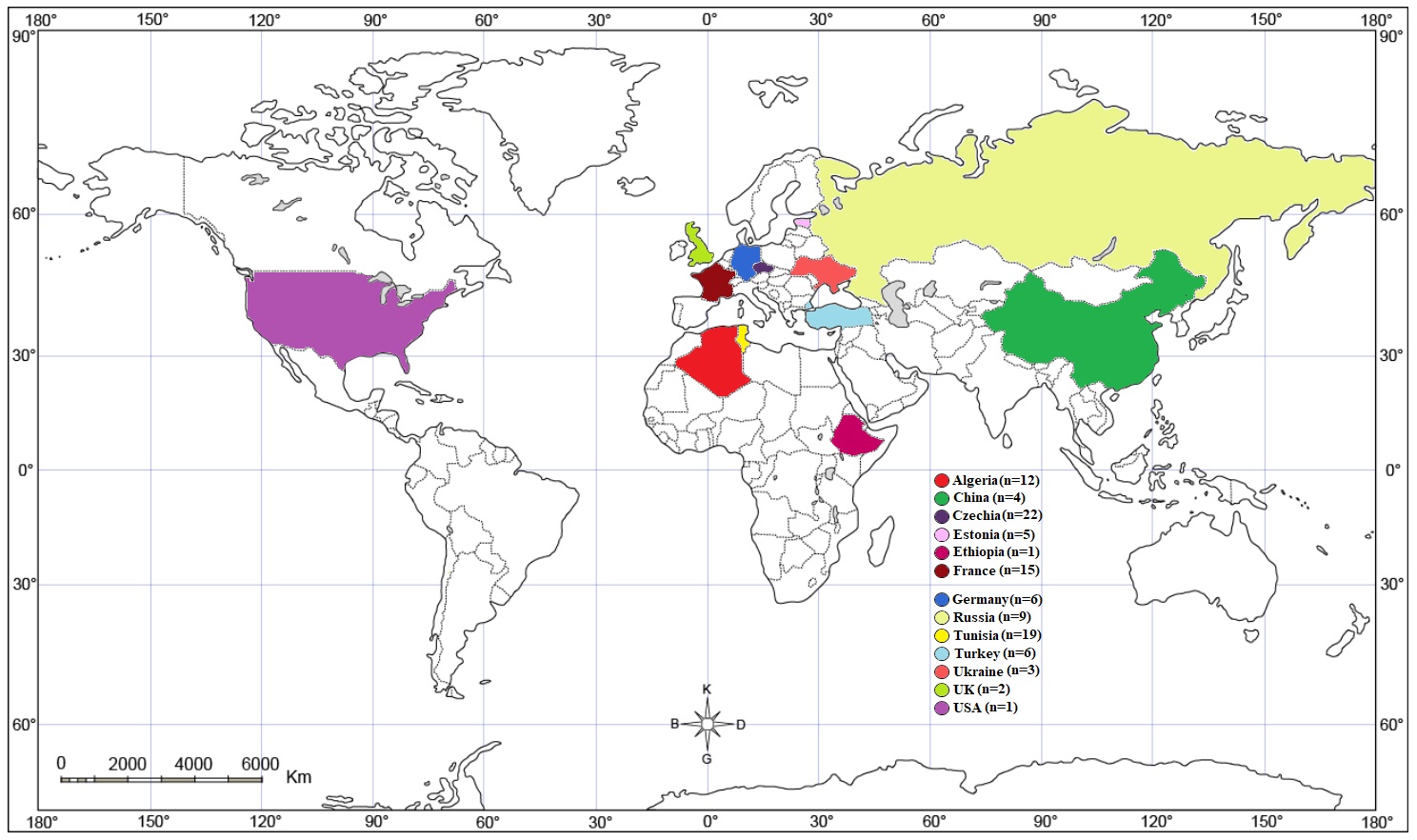
In this study, the geographic origin, fish hosts, and GenBank accession numbers of the mt-CO1 sequences (371 bp) of L. intestinalis isolates were obtained from the NCBI database (Table 1). From NCBI database, 105 mt-CO1 gene sequences (371 bp) were obtained from 13 countries including Algeria (n=12), China (n=4), Czechia (n=22), Estonia (n=5), Ethiopia (n=1), France (n=15), Germany (n=6), Russia (n=9), Tunisia (n=19), Turkey (n=6), Ukraine (n=3), England (n=2) and USA (n=1). The distribution of the sequence-collected countries are shown in Figure 1.
3.1. Diversity, neutrality, fixation and gene flow analysis
Table 2 illustrates the diversity and neutrality indices. The 105 mt-CO1 nucleotide sequences (371 bp) of L. intestinalis isolates from the NCBI database were used to investigate the haplotype and genetic analysis. The resulting sequences lacked appropriately conserved DNA regions. The highest Fixation index (Fst) (0.59761) was found between China and Turkey, while the lowest (-0.10526) was found between Russia and Turkey (Table 3). A negative Tajima's D means a lot of low-frequency polymorphism. In two cases, we encountered a negative Fu’s Fs value, either as evidence for a large number of alleles, as would be expected from a recent population increase or genetic hitchhiking.
Diversity and neutrality indices obtained using nucleotide data of the Ligula intestinalis mt-CO1 gene (371 bp).
Dual fixation index (Fst) for Ligula intestinalis isolates compared with those from various geographic regions using nucleotide data of the mt-CO1 gene.
3.2. Phylogenetic analysis
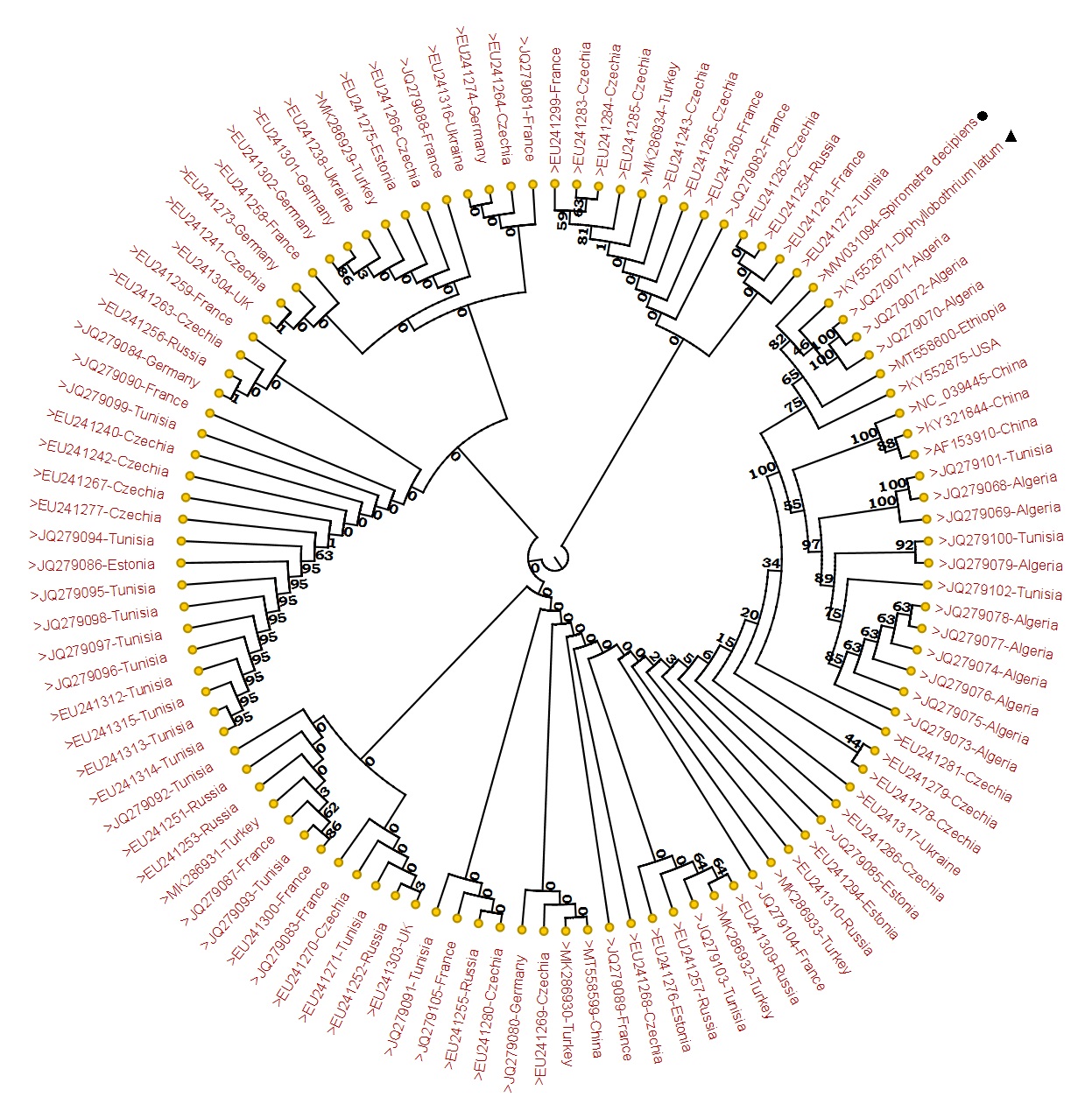
In this study, 105 sequences retrieved from mt-CO1 gene of L. intestinalis isolates were used. Diphyllobothrium latum and Spirometra decipiens sequences were used as outgroup sequences. Figure 2 depicts the phylogenetic tree. Haplotypes were seperated from themselves by mutation at 1-81 different points among themselves. Hap38 (JQ279072-71-70), both Hap35 (JQ279078) and Hap36 (JQ279077-76-75-74) were the most distant haplotypes, showing mutations at 81 points (Table S1).
Phylogenetic tree view of Ligula intestinalis sequences using mt-CO1 gene (371 bp) sequences and reference sequences. CLC Sequence Viewer 8 was used to generate a Maximum Likelihood tree based on the Neighbor Joining model. The reliability of the tree was evaluated with 1000 bootstrap iterations. Spirometra decipiens, Diphyllobothrium latum.
3.3. Haplotype networks
Figure 3 illustrates the haplotype network obtained. Analyses of 38 haplotypes revealed high haplotype diversity (0.791±0.041). L. intestinalis isolates grouping of haplotypes of the mt-CO1 sequences and the accession numbers of the isolates forming the groups are shown in Table S2. The haplotype network obtained as a result of haplotype analysis constitutes a total of 44.76% of Hap24. This haplotype was consisted of 83.33% of Turkey, 80% of France, 77.77% of Russia, 54.54% of Czechia, 50% of Germany, 40% of Estonia, 33.33% of Ukraine and 26.31% of Tunisia sequences. In the haplotype network, 73.68% were single haplotype, 13.15% were double haplotype, 5.26% were triple haplotype, 2.63% were quadruple haplotype, and 2.63% were decimal haplotype.
Appearance of mt-CO1 (371 bp) haplotypes of Ligula intestinalis sequences. The number of mutations that distinguish haplotypes are indicated by screening marks. The geographical distribution of haplotypes is shown in different colors. The size of the circles is related to the haplotype frequency.
4. Discussion
Due to the rapidly increasing population in the world, the need for food has become one of the most important problems. Especially fish are among the most important because they are rich in protein, economically cheap and easily accessible (Oktener et al., 2008OKTENER, A., EGRIBAS, E. and BASUSTA, N., 2008. A preliminary investigation on serious mortalities of fish in Balıklıgöl (Halil-ür Rahman Gölü, Şanlıurfa). Gazi University Journal of Science, vol. 21, no. 1, pp. 9-13.). As aquaculture has become important, aquaculture diseases that develop due to various factors such as bacterial, fungal and viral, etc. have become important today. Among these factors, parasitic diseases constitute the most important part (Oktener, 2003OKTENER, A., 2003. A checklist of metazoan parasites recorded in freshwater fish from Turkey. Zootaxa, vol. 394, no. 1, pp. 1-28. http://dx.doi.org/10.11646/zootaxa.394.1.1.
http://dx.doi.org/10.11646/zootaxa.394.1...
). Therefore, in our study, it was determined that L. intestinalis isolates, which frequently infect fish, it is aimed to determine gene flows, genetic variations and phylogeny according to geographical regions by obtaining 371-bp long sequences belonging to the mt-CO1 gene region.
Mitochondrial DNA sequences are one of the most accurate molecular detection marker for the organism differentiation. It is due to higher mutation rates and maternal inheritance, it is feasible to investigate the genetic analysis of population and their systematic studies of closely related species (Jia et al., 2012JIA, W., YAN, H., LOU, Z., NI, X., DYACHENKO, V., LI, H. and LITTLEWOOD, D.T.J., 2012. Mitochondrial genes and genomes support a cryptic species of tapeworm within Taenia taeniaeformis. Acta Tropica, vol. 123, no. 3, pp. 154-163. http://dx.doi.org/10.1016/j.actatropica.2012.04.006. PMid:22569565.
http://dx.doi.org/10.1016/j.actatropica....
). Sequence data of mt-CO1 gene can be used to investigate phylogenetic relationships at the genus level and to illuminate systematic biology (Chontananarth et al., 2014CHONTANANARTH, T., WONGSAWAD, C., CHOMDEJ, S., KRAILAS, D. and CHAI, J.Y., 2014. Molecular phylogeny of trematodes in family Heterophyidae based on mitochondrial cytochrome c oxidase subunit I (mCOI). Asian Pacific Journal of Tropical Medicine, vol. 7, no. 6, pp. 446-450. http://dx.doi.org/10.1016/S1995-7645(14)60072-9. PMid:25066392.
http://dx.doi.org/10.1016/S1995-7645(14)...
).
Ligula intestinalis is one of the most common tapeworms among freshwater fish and is very common in freshwater basins worldwide. Seasonal migration of their final host waterfowl is very effective in this spread. With the in-silico analysis method, the mt-CO1 gene sequences of L. intestinalis were obtained according to the geographical regions, and the differences in gene sequences were tried to be found by controlling the close relatedness between these isolates and its subspecies have been checked and at the same time, the presence of a gene flow between different geographies was checked.
Neutrality tests such as Tajima D, Fu’s Fs were performed to analyze the neutrality values e.g.h as nucleotide variation and population expansion (Korneliussen et al., 2013KORNELIUSSEN, T.S., MOLTKE, I., ALBRECHTSEN, A. and NIELSEN, R., 2013. Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinformatics, vol. 14, no. 1, p. 289. http://dx.doi.org/10.1186/1471-2105-14-289. PMid:24088262.
http://dx.doi.org/10.1186/1471-2105-14-2...
). While Tajima D based on previous mutations reflecting population events for a long time. Fu’s Fs value is more specific to latest mutation. The sequences of 13 different geographic populations were analyzed reporting low and negative values of Fu's F and Tajima D values. A negative Tajima's D means a lot of low-frequency polymorphism. In two cases, we encounter a negative Fu’s Fs value, either as evidence for a large number of alleles, as would be expected from a recent population increase or genetic hitchhiking. It was seen that the main haplotype obtained in haplotype analysis made up 44.76% of the total network and included isolates from eight different countries. Therefore, this main haplotype represents a single ancestor. When the main haplotype and other haplotypes were analyzed, it was seen that L. intestinalis isolates exhibited a rapid genetic diversity and could lead to the formation of a subspecies.
China and Turkey showed highest fixation index (Fst) valeu of 0.59761, while the lowest (-0.10526) was found between Russia and Turkey. In this scenario, it can be stated that gen flow between the Chinese and Turkish populations is at a minimum level. It may be an indication that there is little or no fish trade, presumably because of China and Turkey are located by the sea in both countries.
As a result of haplotype analysis, 38 haplotypes were formed among 13 different countries. In these, 28 haplotypes were detected as single haplotype. This situation makes us think that it may be an indication that different subspecies may emerge as a result of the evolutionary period.
Haplotypes were separated from themselves by mutation at 1-81 different points among themselves. Hap38 (JQ279072-71-70), both Hap35 (JQ279078) and Hap36 (JQ279077-76-75-74) were the most distant haplotypes, showing mutations at 81 points. This situation made us think that there may be a gene flow as a result of cross fertilization between the species brought from Europe to North Africa, since the city of Algeria has a coastline to the Mediterranean Sea and is on the migration routes of waterfowl.
5. Conclusion
In conclusion, there have been different molecular studies about L. intestinalis carried out to date, but it is the first study done as an in-silico analysis method. With this study, it serves as a stepping-stone for future studies on L. intestinalis worldwide on epidemiology, genetic variation, and evolutionary process. The mt-CO1 sequences sent to the dataset at NCBI included regions of short length, which were the most important limiting factors for us. Therefore, in future studies, it will be useful to target larger gene regions and genetic diversity in more comprehensive and to analyze them in terms of possible new subspecies.
Supplementary Material
Supplementary material accompanies this paper.
Table S1 Nucleotide variation positions of the mt-CO1 (371 bp) gene among 38 haplotypes analyzed. Table S2 Haplotype of mt-CO1 sequences of Ligula intestinalis and accession numbers of isolates forming groups.This material is available as part of the online article from https://www.scielo.br/j/bjb
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81971969 and 81772225 to JC), and the Three-Year Public Health Action Plan of Shanghai (2020-2022), China (No. GWV-10.1-XK13 to JC). The funders had no role in the study design, the data collection and analysis, the decision to publish, or the preparation of the manuscript.
References
- ALI, M.A., GYULAI, G., HIDVÉGI, N., KERTI, B., HEMAID, F.M., PANDEY, A.K. and LEE, J., 2014. The changing epitome of species identification–DNA barcoding. Saudi Journal of Biological Sciences, vol. 21, no. 3, pp. 204-231. http://dx.doi.org/10.1016/j.sjbs.2014.03.003 PMid:24955007.
» http://dx.doi.org/10.1016/j.sjbs.2014.03.003 - BARBER, I., HOARE, D. and KRAUSE, J., 2000. Effects of parasites on fish behaviour: a review and evolutionary perspective. Reviews in Fish Biology and Fisheries, vol. 10, no. 2, pp. 131-165. http://dx.doi.org/10.1023/A:1016658224470
» http://dx.doi.org/10.1023/A:1016658224470 - BARSON, M. and MARSHALL, B., 2003. The occurrence of the tapeworm Ligula intestinalis (L.), in Barbus paludinosus from a small dam in Zimbabwe. African Journal of Aquatic Science, vol. 28, no. 2, pp. 175-178. http://dx.doi.org/10.2989/16085910309503782
» http://dx.doi.org/10.2989/16085910309503782 - BRITTON, J.R., JACKSON, M.C. and HARPER, D.M., 2009. Ligula intestinalis (Cestoda: Diphyllobothriidae) in Kenya: a field investigation into host specificity and behavioural alterations. Parasitology, vol. 136, no. 11, pp. 1367-1373. http://dx.doi.org/10.1017/S003118200999059X PMid:19627635.
» http://dx.doi.org/10.1017/S003118200999059X - CHONTANANARTH, T., WONGSAWAD, C., CHOMDEJ, S., KRAILAS, D. and CHAI, J.Y., 2014. Molecular phylogeny of trematodes in family Heterophyidae based on mitochondrial cytochrome c oxidase subunit I (mCOI). Asian Pacific Journal of Tropical Medicine, vol. 7, no. 6, pp. 446-450. http://dx.doi.org/10.1016/S1995-7645(14)60072-9 PMid:25066392.
» http://dx.doi.org/10.1016/S1995-7645(14)60072-9 - EMAMINEW, T., DEREJE, B. and ABDU, M., 2014. Prevalence of Ligula intestinalis larvae in Barbus fish genera at Lake Tana, Ethiopia. World Journal of Fish and Marine Sciences, vol. 6, no. 5, pp. 408-416.
- GABAGAMBI, N.P. and SKORPING, A., 2018. Spatial and temporal distribution of Ligula intestinalis (Cestoda: Diphyllobothriidea) in usipa (Engraulicypris sardella)(Pisces: Cyprinidae) in Lake Nyasa. Journal of Helminthology, vol. 92, no. 4, pp. 410-416. http://dx.doi.org/10.1017/S0022149X17000724 PMid:28818117.
» http://dx.doi.org/10.1017/S0022149X17000724 - HAJIBABAEI, M., SINGER, G.A., HEBERT, P.D. and HICKEY, D.A., 2007. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends in Genetics, vol. 23, no. 4, pp. 167-172. http://dx.doi.org/10.1016/j.tig.2007.02.001 PMid:17316886.
» http://dx.doi.org/10.1016/j.tig.2007.02.001 - HEBERT, P.D., CYWINSKA, A., BALL, S.L. and DEWAARD, J.R., 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B, vol. 270, no. 1512, pp. 313-321. http://dx.doi.org/10.1098/rspb.2002.2218 PMid:12614582.
» http://dx.doi.org/10.1098/rspb.2002.2218 - HOOLE, D., CARTER, V. and DUFOUR, S., 2010. Ligula intestinalis (cestoda: Pseudophyllidea): an ideal fish-metazoan parasite model? Parasitology, vol. 137, no. 3, pp. 425-438. http://dx.doi.org/10.1017/S0031182010000107 PMid:20163752.
» http://dx.doi.org/10.1017/S0031182010000107 - JIA, W., YAN, H., LOU, Z., NI, X., DYACHENKO, V., LI, H. and LITTLEWOOD, D.T.J., 2012. Mitochondrial genes and genomes support a cryptic species of tapeworm within Taenia taeniaeformis. Acta Tropica, vol. 123, no. 3, pp. 154-163. http://dx.doi.org/10.1016/j.actatropica.2012.04.006 PMid:22569565.
» http://dx.doi.org/10.1016/j.actatropica.2012.04.006 - KOC, H.T., ERDOGAN, Z. and COZ-RAKOVAC, R., 2006. The occurrence of Ligula intestinalis (L.) observed in chub (Leuciscus cephalus L.) from Caparlipatlak Dam lake, Ivrindi-Balikesir, Turkey. Periodicum Biologorum, vol. 108, no. 2, pp. 183-187.
- KORNELIUSSEN, T.S., MOLTKE, I., ALBRECHTSEN, A. and NIELSEN, R., 2013. Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinformatics, vol. 14, no. 1, p. 289. http://dx.doi.org/10.1186/1471-2105-14-289 PMid:24088262.
» http://dx.doi.org/10.1186/1471-2105-14-289 - LEIGH, J.W. and BRYANT, D., 2015. Popart: full‐feature software for haplotype network construction. Methods in Ecology and Evolution, vol. 6, no. 9, pp. 1110-1116. http://dx.doi.org/10.1111/2041-210X.12410
» http://dx.doi.org/10.1111/2041-210X.12410 - LOOT, G., AULAGNIER, S., LEK, S., THOMAS, F. and GUÉGAN, J.F., 2002. Experimental demonstration of a behavioural modification in a cyprinid fish, Rutilus rutilus (L.), induced by a parasite, Ligula intestinalis (L.). Canadian Journal of Zoology, vol. 80, no. 4, pp. 738-744. http://dx.doi.org/10.1139/z02-043
» http://dx.doi.org/10.1139/z02-043 - LOOT, G., FRANCISCO, P., SANTOUL, F., LEK, S. and GUÉGAN, J.F., 2001. The three hosts of the Ligula intestinalis (Cestoda) life cycle in Lavernose-Lacasse gravel pit, France. Archiv für Hydrobiologie, vol. 152, no. 3, pp. 511-525. http://dx.doi.org/10.1127/archiv-hydrobiol/152/2001/511
» http://dx.doi.org/10.1127/archiv-hydrobiol/152/2001/511 - MADDISON, D.R., SWOFFORD, D.L. and MADDISON, W.P., 1997. NEXUS: an extensible file format for systematic information. Systematic Biology, vol. 46, no. 4, pp. 590-621. http://dx.doi.org/10.1093/sysbio/46.4.590 PMid:11975335.
» http://dx.doi.org/10.1093/sysbio/46.4.590 - OKTENER, A., 2003. A checklist of metazoan parasites recorded in freshwater fish from Turkey. Zootaxa, vol. 394, no. 1, pp. 1-28. http://dx.doi.org/10.11646/zootaxa.394.1.1
» http://dx.doi.org/10.11646/zootaxa.394.1.1 - OKTENER, A., EGRIBAS, E. and BASUSTA, N., 2008. A preliminary investigation on serious mortalities of fish in Balıklıgöl (Halil-ür Rahman Gölü, Şanlıurfa). Gazi University Journal of Science, vol. 21, no. 1, pp. 9-13.
- OSUIGWE, D. and OBIEKEZIE, A., 2007. Assessment of the growth performance and feed utilization of fingering Heterobranchus longifilis fed raw and boiled jackbean (Canavalia ensiformis) seed meal as fishmeal substitute. Journal of Fisheries International, vol. 2, pp. 37-41.
- OZBEK, M. and OZTURK, M.O., 2010. Investigations on Ligula intestinalis plerocercoid L., 1758 infection of some fishes from Dam lake Kunduzlar (Kirka, Eskişehir). Turkiye Parazitoloji Dergisi, vol. 34, no. 2, pp. 112-117. PMid:20597057.
- ROZAS, J., FERRER-MATA, A., SÁNCHEZ-DELBARRIO, J.C., GUIRAO-RICO, S., LIBRADO, P., RAMOS-ONSINS, S.E. and SÁNCHEZ-GRACIA, A., 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, vol. 34, no. 12, pp. 3299-3302. http://dx.doi.org/10.1093/molbev/msx248 PMid:29029172.
» http://dx.doi.org/10.1093/molbev/msx248 - SARKAR, I.N. and TRIZNA, M., 2011. The Barcode of Life Data Portal: bridging the biodiversity informatics divide for DNA barcoding. PLoS One, vol. 6, no. 7, p. e14689. http://dx.doi.org/10.1371/journal.pone.0014689 PMid:21818249.
» http://dx.doi.org/10.1371/journal.pone.0014689 - SHARGH, S., SHAMSAII, M. and KARIMI, S., 2008. Distribution of parasitic Cestod “Ligula intestinalis” in Mazandaran region. Iranian Journal of Parasitology, vol. 3, no. 2, pp. 26-33.
- TIBAYRENC, M., 2005. Bridging the gap between molecular epidemiologists and evolutionists. Trends in Microbiology, vol. 13, no. 12, pp. 575-580. http://dx.doi.org/10.1016/j.tim.2005.09.004 PMid:16214342.
» http://dx.doi.org/10.1016/j.tim.2005.09.004 - TURGUT, E., DEVELI, N., YESILAYER, N. and BUHAN, E., 2011. Seasonal occurrence of Ligula intestinalis infection in cyprinids from Almus Dam Lake (Turkey). KSÜ Doğa Bilimleri Dergisi, vol. 14, no. 3, pp. 9-11.
- URDES, L.D. and HANGAN, M., 2013. The epidemiology of Ligula intestinalis (Phylum Platyhelminthes) within the cyprinid populations inhabiting the Danubian Delta area. Scientific Papers Animal Science and Biotechnologies, vol. 46, no. 1, pp. 273-276.
- WELIANGE, W. and AMARASINGHE, U., 2001. The occurrence of cestode Ligula intestinalis (Linnaeus) from attentive carplet Amblypharyngodon melettinus (Valenciennes) in Sri Lanka. Asian Fisheries Science, vol. 14, no. 1, pp. 95-98. http://dx.doi.org/10.33997/j.afs.2001.14.1.011
» http://dx.doi.org/10.33997/j.afs.2001.14.1.011 - QIAGEN CLC MAIN WORKBENCH, 2007. QIAGEN CLC Main Workbench: the user-friendly solution for basic sequencing analysis [software]. Hilden: QIAGEN.
Publication Dates
-
Publication in this collection
10 June 2022 -
Date of issue
2024
History
-
Received
27 Nov 2021 -
Accepted
01 Mar 2022