Abstract
We studied the complete chloroplast genome of Gomphocarpus siniacus and Duvalia velutina from Asclepiadoideae subfamily; due to their medicinal importance and distribution worldwide their interest became high. In this study we analyzed the complete chloroplast genomes of G. siniacus and D. velutina using Illumina sequencing technology. The sequences were compared with the other species from Apocynaceae family. The complete genome of G. siniacus is 162,570 bp while D. velutina has154, 478 bp in length. Both genomes consist of 119 genes; encode 31 tRNA genes, and eight rRNA genes. Comparative studies of the two genomes showed variations in SSR markers in which G. siniacus possesses 223 while D. velutina has 186. This could be used for barcoding in order to aid in easy identification of the species. Phylogenetic analysis on the other hand reaffirms the tribal position of G. siniacus in Asclepiadeae and D. velutina in Ceropegieae. These findings could be used in subsequent research studies of angiosperms identification, genetic engineering, herb genomics and phylogenomic studies of Apocynaceae family.
Keywords:
chloroplast genomics; Duvalia velutina; evolution; Gomphocarpus siniacus; phylogenetic tree
Resumo
Estudamos o genoma completo do cloroplasto de Gomphocarpus siniacus e Duvalia velutina da subfamília Asclepiadoideae. Em razão de sua importância medicinal e distribuição em todo o mundo, o seu interesse tornou-se elevado. Neste estudo, analisamos os genomas completos de cloroplastos de G. siniacus e D. velutina usando a tecnologia de sequenciamento Illumina. As sequências foram comparadas com as demais espécies da família Apocynaceae. O genoma completo de G. siniacus tem 162.570 pb, enquanto D. velutina tem 154.478 pb de comprimento. Ambos os genomas consistem em 119 genes e codificam 31 genes de tRNA e 8 genes de rRNA. Estudos comparativos dos dois genomas mostraram variações nos marcadores SSR em que G. siniacus possui 223, enquanto D. velutina possui 186. Isso poderia ser usado para código de barras para facilitar a identificação das espécies. A análise filogenética, por outro lado, reafirma a posição tribal de G. siniacus em Asclepiadeae e D. velutina em Ceropegieae. Esses achados poderão ser utilizados em pesquisas posteriores de identificação de angiospermas, engenharia genética, genômica de ervas e estudos filogenômicos da família Apocynaceae.
Palavras-chave:
genômica de cloroplastos; Duvalia velutina; evolução; Gomphocarpus siniacus; árvore filogenética
1. Introduction
Duvalia velutina (Lavranos) is a succulent, perennial plant distributed in Saudi Arabia, South Africa, Yemen and Namibia. It had distinct floral and stem characteristics which are common in almost all the species as such can only be distinguished when flowers appeared. D. velutina usually grow in colonies and it is locally used as food and medicine as well as ornamental purposes (Burkill, 2004BURKILL, H.M., 2004. The useful plants of West tropical Africa. Kew: Royal Botanic Gardens, vol. 6.). Gomphocarpus siniacus (Boiss) is an herbaceous plant which is found in Africa, Saudi Arabia, Egypt (Sinai Peninsula), Jordan, and Yemen. In Africa, G. siniacus is used for several medicinal and other uses, such as treatment of bile, tuberculosis and stomach pain. Many active compounds have been reported in these plants for example in G. siniacus contains some cardio-active glycosides and alkaloids (Burkill, 2004BURKILL, H.M., 2004. The useful plants of West tropical Africa. Kew: Royal Botanic Gardens, vol. 6.). Modern pharmacological investigations also showed their possible anti oxidative and antibacterial potentials. Similarly, D. velutina and G. siniacus belongs to subfamily Asclepiadoideae in the family Apocynaceae. Although previous studies on D. velutina and G. siniacus focuses on eco-physiology with few studies on molecular genetics (Lang et al., 2018LANG, Y., WANG, M., XIA, J. and ZHAO, Q., 2018. Effects of soil drought stress on photosynthetic gas exchange traits and chlorophyll fluorescence in Forsythia suspensa. Journal of Forestry Research, vol. 29, no. 1, pp. 45-53. http://dx.doi.org/10.1007/s11676-017-0420-9.
http://dx.doi.org/10.1007/s11676-017-042...
; Masrahi, 2015MASRAHI, Y.S., 2015. A new species of Leptadenia (Apocynaceae) and two other new records from southwestern Saudi Arabia. Saudi Journal of Biological Sciences, vol. 22, no. 5, pp. 631-636. http://dx.doi.org/10.1016/j.sjbs.2015.02.003. PMid:26288569.
http://dx.doi.org/10.1016/j.sjbs.2015.02...
). Also, so many attentions have been given to cultivated variety of D. velutina and G. siniacus as a result the wild varieties has been neglected; among the Gomphocarpus sp. only G. siniacus is native to Arabian Peninsula therefore it is very important for conservation and pharmaceutical discovery as well as the evolutionary studies. Majority of Asclepiadoideae species do not have a common character unique to their clades which results in wrong identifications (Abba et al., 2020ABBA, A., ALZAHRANI, D., YARADUA, S. and ALBOKHARI, E.B., 2020. Complete plastome genome of Pergularia tomentosa L. (Asclepiadoideae, Apocynaceae). Mitochondrial DNA. Part B, Resources, vol. 5, no. 1, pp. 566-567. http://dx.doi.org/10.1080/23802359.2019.1710291. PMid:33366649.
http://dx.doi.org/10.1080/23802359.2019....
).
There is need for effective molecular markers to address problem of identification in Asclepiadoideae. Some of the markers used to identify D. velutina and G. siniacus were ITS, psbA, matK and rbcL which has helped to some extent in the recent taxonomy of Apocynaceae (Masrahi, 2015MASRAHI, Y.S., 2015. A new species of Leptadenia (Apocynaceae) and two other new records from southwestern Saudi Arabia. Saudi Journal of Biological Sciences, vol. 22, no. 5, pp. 631-636. http://dx.doi.org/10.1016/j.sjbs.2015.02.003. PMid:26288569.
http://dx.doi.org/10.1016/j.sjbs.2015.02...
). Due to the important economic and medicinal benefits of these species there have been adulterants stocks being sold to people in place of the original species which can affect the quality and medicinal efficacy of the target drugs and or concoctions. For this reason a correct identification of these species will greatly protect the genetic resources and lineage history. The widely used method of identifying these species was ITS and multiple genomic partial segment bar-coding (Masrahi, 2015MASRAHI, Y.S., 2015. A new species of Leptadenia (Apocynaceae) and two other new records from southwestern Saudi Arabia. Saudi Journal of Biological Sciences, vol. 22, no. 5, pp. 631-636. http://dx.doi.org/10.1016/j.sjbs.2015.02.003. PMid:26288569.
http://dx.doi.org/10.1016/j.sjbs.2015.02...
; Lang et al., 2018LANG, Y., WANG, M., XIA, J. and ZHAO, Q., 2018. Effects of soil drought stress on photosynthetic gas exchange traits and chlorophyll fluorescence in Forsythia suspensa. Journal of Forestry Research, vol. 29, no. 1, pp. 45-53. http://dx.doi.org/10.1007/s11676-017-0420-9.
http://dx.doi.org/10.1007/s11676-017-042...
). The commonly used plastid regions were PsbA, trnH and matK are not enough in the correct identification of some angiosperms (Cui et al., 2019CUI, Y., ZHOU, J., CHEN, X., XU, Z., WANG, Y., SUN, W., SONG, J. and YAO, H., 2019. Complete chloroplast genome and comparative analysis of three Lycium (Solanaceae) species with medicinal and edible properties. Gene Reports, vol. 17, p. 100464. http://dx.doi.org/10.1016/j.genrep.2019.100464.
http://dx.doi.org/10.1016/j.genrep.2019....
). Chloroplast as one of the major differences between plant cells and animal cell plays a crucial role in providing energy for plants metabolism (Li et al., 2013LI, X., ZHANG, T.-C., QIAO, Q., REN, Z., ZHAO, J., YONEZAWA, T., HASEGAWA, M., CRABBE, M.J.C., LI, J. and ZHONG, Y., 2013. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS One, vol. 8, no. 3, p. e58747. http://dx.doi.org/10.1371/journal.pone.0058747. PMid:23554920.
http://dx.doi.org/10.1371/journal.pone.0...
; Neuhaus and Emes, 2000NEUHAUS, H.E. and EMES, M.J., 2000. Nonphotosynthetic metabolism in plastids. Annual Review of Plant Physiology and Plant Molecular Biology, vol. 51, no. 1, pp. 111-140. http://dx.doi.org/10.1146/annurev.arplant.51.1.111. PMid:15012188.
http://dx.doi.org/10.1146/annurev.arplan...
; Rodríguez-Ezpeleta et al., 2005RODRÍGUEZ-EZPELETA, N., BRINKMANN, H., BUREY, S.C., ROURE, B., BURGER, G., LÖFFELHARDT, W., BOHNERT, H.J., PHILIPPE, H. and LANG, B.F., 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Current Biology, vol. 15, no. 14, pp. 1325-1330. http://dx.doi.org/10.1016/j.cub.2005.06.040. PMid:16051178.
http://dx.doi.org/10.1016/j.cub.2005.06....
). For many decades and now chloroplast genomes have been given many attention by plant researchers. Quadripartite structure is the main symbol of chloroplast genome and a 115 kb – 165 kb sequence length with large single-copy region (LSC), a small single-copy region (SSC) and a pair of inverted repeats regions IRa and IRb (Dong et al., 2012DONG, W., LIU, J., YU, J., WANG, L. and ZHOU, S., 2012. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One, vol. 7, no. 4, p. e35071. http://dx.doi.org/10.1371/journal.pone.0035071. PMid:22511980.
http://dx.doi.org/10.1371/journal.pone.0...
; Jansen et al., 2005JANSEN, J.J.P., VAN DEN BOSCH, F.A.J. and VOLBERDA, H.W., 2005. Managing potential and realized absorptive capacity: how do organizational antecedents matter? Academy of Management Journal, vol. 48, no. 6, pp. 999-1015. http://dx.doi.org/10.5465/amj.2005.19573106.
http://dx.doi.org/10.5465/amj.2005.19573...
). Chloroplast (cp) genome is very conserved in terms of its gene order and contents compared to other organelles in majority of angiosperms (Tonti‐Filippini et al., 2017TONTI‐FILIPPINI, J., NEVILL, P.G., DIXON, K. and SMALL, I., 2017. What can we do with 1000 plastid genomes? The Plant Journal, vol. 90, no. 4, pp. 808-818. http://dx.doi.org/10.1111/tpj.13491. PMid:28112435.
http://dx.doi.org/10.1111/tpj.13491...
; Wicke et al., 2011WICKE, S., SCHNEEWEISS, G.M., DEPAMPHILIS, C.W., MÜLLER, K.F. and QUANDT, D., 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Molecular Biology, vol. 76, no. 3-5, pp. 273-297. http://dx.doi.org/10.1007/s11103-011-9762-4. PMid:21424877.
http://dx.doi.org/10.1007/s11103-011-976...
). With that reason the evolutionary history details of most angiosperms express a unique and vital information for plant phylogeny (Corriveau and Coleman, 1988CORRIVEAU, J.L. and COLEMAN, A.W., 1988. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany, vol. 75, no. 10, pp. 1443-1458. http://dx.doi.org/10.1002/j.1537-2197.1988.tb11219.x.
http://dx.doi.org/10.1002/j.1537-2197.19...
). Several research on DNA-bar-coding as well as the molecular markers give more emphasis on the chloroplast genome after the work Nicotina tabacum by Shinozaki et al. (1986)SHINOZAKI, K., OHME, M., TANAKA, M., WAKASUGI, T., HAYASHIDA, N., MATSUBAYASHI, T., ZAITA, N., CHUNWONGSE, J., OBOKATA, J., YAMAGUCHI‐SHINOZAKI, K., OHTO, C., TORAZAWA, K., MENG, B.Y., SUGITA, M., DENO, H., KAMOGASHIRA, T., YAMADA, K., KUSUDA, J., TAKAIWA, F., KATO, A., TOHDOH, N., SHIMADA, H. and SUGIURA, M., 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. The EMBO Journal, vol. 5, no. 9, pp. 2043-2049. http://dx.doi.org/10.1002/j.1460-2075.1986.tb04464.x. PMid:16453699.
http://dx.doi.org/10.1002/j.1460-2075.19...
. Many plant biologist were able to identify over 3000 plant chloroplast genomes and stored in the GenBank (Mocan et al., 2014MOCAN, A., VLASE, L., VODNAR, D.C., BISCHIN, C., HANGANU, D., GHELDIU, A.-M., OPREAN, R., SILAGHI-DUMITRESCU, R. and CRIȘAN, G., 2014. Polyphenolic content, antioxidant and antimicrobial activities of Lycium barbarum L. and Lycium chinense Mill. leaves. Molecules, vol. 19, no. 7, pp. 10056-10073. http://dx.doi.org/10.3390/molecules190710056. PMid:25014533.
http://dx.doi.org/10.3390/molecules19071...
; NCBI). Moreover, the comparative chloroplast genomes of two Asclepiadoideae species will reveal the Phylogenetic lineage between the two species and their positions in Asclepiadoideae well as the tribal positions.
The characterization of highly variable regions would contribute to developing candidate DNA barcodes for future studies. Microsatellites (SSRs) could be used as potential molecular polymorphic markers to reveal the genetic diversity and population structure of Apocynaceae. The detection of protein-coding genes under intense selection pressure could play an important role in the analyses of evolution and adaptation of plants in an ecosystem. in addition, this study would reconstruct the intergeneric relationships and locate the phylogenetic position of sub family Asclepiadoideae.
2. Materials and Methods
2.1. Plant sampling, DNA extractions and sequencing
Fresh leaves of D. velutina and G. siniacus were obtained from the Ash-shafa Mountains in At-Taif city of Makkah Region, Saudi Arabia (21◦ 4.7’33’N; 41◦17.9’ 29” E) on the 2nd of June 2019. Samples were identified at the herbarium of Department of Biological Sciences, Faculty of Science of King Abdulaziz University Jeddah KSA. The voucher specimens of the two plants species were deposited at the herbarium of King Abdulaziz University, Jeddah. The leaves were washed with 70% ethanol and then DNA was extracted using DNeasy Plant mini kit following standard protocol (Qiagen Co. Germany). Quality of the DNA was checked using Nanodrop 2000C Spectrophotometer and Electrophoresis in 1% (w/v) agarose gel. The pure DNA was used to construct the libraries and was sequenced with Illumina Hiseq 2500 (Beijing, China) following the standard protocol. The result of 6.8 Gb of D. velutina and 7.1 G. siniacus and pair reads of 500 bp were recovered.
2.2. Chloroplast genome assembly and annotation
The Raw sequence was filtered using Skewer 0.2.2 and trimmed with Trimmomatic V).36 (Bolger et al., 2014BOLGER, A.M., LOHSE, M. and USADEL, B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, vol. 30, no. 15, pp. 2114-2120. http://dx.doi.org/10.1093/bioinformatics/btu170. PMid:24695404.
http://dx.doi.org/10.1093/bioinformatics...
). Sequences were mapped with the reference genome from the NCBI using BLASTN with default settings. In which the Stapelia gigantea (MG963259) complete chloroplast genome was used as reference to assemble the D. velutina while Cynanchum wilfordii (KT220734) was used to assemble the G. siniacus genome. SOAPdenovo (Xie et al., 2014XIE, Y., WU, G., TANG, J., LUO, R., PATTERSON, J., LIU, S., HUANG, W., HE, G., GU, S., LI, S., ZHOU, X., LAM, T.-W., LI, Y., XU, X., WONG, G.K.-S. and WANG, J., 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics, vol. 30, no. 12, pp. 1660-1666. http://dx.doi.org/10.1093/bioinformatics/btu077. PMid:24532719.
http://dx.doi.org/10.1093/bioinformatics...
) was applied to assemble the contigs while the annotation was done using PGA software (Qu et al., 2019QU, X.-J., MOORE, M.J., LI, D.-Z. and YI, T.-S., 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, vol. 15, no. 1, p. 50. http://dx.doi.org/10.1186/s13007-019-0435-7. PMid:31139240.
http://dx.doi.org/10.1186/s13007-019-043...
).
Geseq was used to annotate genes (Lohse et al., 2007LOHSE, M., DRECHSEL, O. and BOCK, R., 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics, vol. 52, no. 5-6, pp. 267-274. http://dx.doi.org/10.1007/s00294-007-0161-y. PMid:17957369.
http://dx.doi.org/10.1007/s00294-007-016...
), while ARAGORN V 1.2.2 and tRNAscan-SE V 2.0.3 was used for the annotation of tRNAs in the sequences (Lowe and Chan, 2016LOWE, T.M. and CHAN, P.P., 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Research, vol. 44, no. W1, pp. W54-W57. http://dx.doi.org/10.1093/nar/gkw413. PMid:27174935.
http://dx.doi.org/10.1093/nar/gkw413...
). For circular genome structure we use Organellar Genome DRAW (Tillich et al., 2017TILLICH, M., LEHWARK, P., PELLIZZER, T., ULBRICHT-JONES, E.S., FISCHER, A., BOCK, R. and GREINER, S., 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Research, vol. 45, no. W1, pp. W6-W11. http://dx.doi.org/10.1093/nar/gkx391. PMid:28486635.
http://dx.doi.org/10.1093/nar/gkx391...
). The annotated sequences of D. velutina and G. siniacus were submitted to National center for Biotechnology Information (NCBI) and were assigned with an accession Numbers MT431578 and MN689141 for D. velutina and G. siniacus respectively.
2.3. Comparative analysis and genomic features
Comparative genomics was made using mVISTA programme (Mayor et al., 2000MAYOR, C., BRUDNO, M., SCHWARTZ, J.R., POLIAKOV, A., RUBIN, E.M., FRAZER, K.A., PACHTER, L.S. and DUBCHAK, I., 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics, vol. 16, no. 11, pp. 1046-1047. http://dx.doi.org/10.1093/bioinformatics/16.11.1046. PMid:11159318.
http://dx.doi.org/10.1093/bioinformatics...
); to compare D. velutina and G. siniacus, sequences with Calotropis procera (NC_041440) and Gymnema sylvestre (NC_047175) genomes both from Asclepiadoideae downloaded from GenBank database. While the expansion and contractions of the sequences were done using IR scope (Amiryousefi et al., 2018AMIRYOUSEFI, A., HYVÖNEN, J. and POCZAI, P., 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics, vol. 34, no. 17, pp. 3030-3031. http://dx.doi.org/10.1093/bioinformatics/bty220. PMid:29659705.
http://dx.doi.org/10.1093/bioinformatics...
).
2.4. Amino acid frequency, codon usage, and RNA editing sites
For the sequence analysis we use MEGA 6.0 (Kumar et al., 2008KUMAR, S., NEI, M., DUDLEY, J. and TAMURA, K., 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics, vol. 9, no. 4, pp. 299-306. http://dx.doi.org/10.1093/bib/bbn017. PMid:18417537.
http://dx.doi.org/10.1093/bib/bbn017...
) to detect the relative synonymous codon usage (RSCU), codon usage as well as the base compositions; while RNA editing sites in the protein coding genes were analyzed with PREP suite (Mower, 2009MOWER, J.P., 2009. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Research, vol. 37, suppl. 2, pp. W253-W259. http://dx.doi.org/10.1093/nar/gkp337. PMid:19433507.
http://dx.doi.org/10.1093/nar/gkp337...
) with 0.8 cutoff values.
2.5. Microsatellites analysis
Microsatellites in the sequences of G. siniacus and D. velutina were evaluated with MISA (IPGCPR, Gatersleben, Germany) (Thiel et al., 2003THIEL, T., MICHALEK, W., VARSHNEY, R. and GRANER, A., 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theoretical and Applied Genetics, vol. 106, no. 3, pp. 411-422. http://dx.doi.org/10.1007/s00122-002-1031-0. PMid:12589540.
http://dx.doi.org/10.1007/s00122-002-103...
) with the settings of 10, 5, 4, 3, 3and 3to represent mono, di tri, tetra, penta and hexa values respectively; while Tandem Repeats were identified with a program called Tandem repeat Finder (NY, USA) with ten base pairs length. While setting 2, 7, 7, for match, mismatch and indels respectively. The size of the repeats were viewed with program REPUter (Beilfeild Germany) (Kurtz et al., 2001KURTZ, S., CHOUDHURI, J.V., OHLEBUSCH, E., SCHLEIERMACHER, C., STOYE, J. and GIEGERICH, R., 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research, vol. 29, no. 22, pp. 4633-4642. http://dx.doi.org/10.1093/nar/29.22.4633. PMid:11713313.
http://dx.doi.org/10.1093/nar/29.22.4633...
) parameters were set at 30 base pairs as least size and ninety percent limit similarity index of two repeat copies.
2.6. Substitutions and InDel analyses
In order to determined substitution rates in D. velutina and G. siniacus, the sequence of Gymnema sylvestre was used as a reference (Yaradua et al., 2019YARADUA, S.S., ALZAHRANI, D.A., ALBOKHARY, E.J., ABBA, A. and BELLO, A., 2019. Complete chloroplast genome sequence of Justicia flava: genome comparative analysis and phylogenetic relationships among Acanthaceae. BioMed Research International, vol. 2019, p. 4370258. http://dx.doi.org/10.1155/2019/4370258. PMid:31467890.
http://dx.doi.org/10.1155/2019/4370258...
) while alignment of SSC, LSC and IR regions in the genomes was done using MAFFTv.5 (Multiple Alignment with Fast Fourier Transform) (Katoh and Standley, 2016KATOH, K. and STANDLEY, D.M., 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics, vol. 32, no. 13, pp. 1933-1942. http://dx.doi.org/10.1093/bioinformatics/btw108. PMid:27153688.
http://dx.doi.org/10.1093/bioinformatics...
). The numbers and types of substitutions were described in Geneious R8.1(Kearse et al., 2012KEARSE, M., MOIR, R., WILSON, A., STONES-HAVAS, S., CHEUNG, M., STURROCK, S., BUXTON, S., COOPER, A., MARKOWITZ, S., DURAN, C., THIERER, T., ASHTON, B., MEINTJES, P. and DRUMMOND, A., 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, vol. 28, no. 12, pp. 1647-1649. http://dx.doi.org/10.1093/bioinformatics/bts199. PMid:22543367.
http://dx.doi.org/10.1093/bioinformatics...
). InDels events were determined after analyzing a pairwise alignment the SSC, LSC and IR in DnaSP v.5.10 (Librado and Rozas, 2009LIBRADO, P. and ROZAS, J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, vol. 25, no. 11, pp. 1451-1452. http://dx.doi.org/10.1093/bioinformatics/btp187. PMid:19346325.
http://dx.doi.org/10.1093/bioinformatics...
).
2.7. Phylogenetic analysis
Complete plastome genome of D. velutina and G. siniacus along with other 9 species from Asclepiadoideae subfamily. Two species were outgroup from Rauvofluideae subfamily; were downloaded from the Genbank and aligned with MAFFT program v.7 (Katoh and Standley, 2013KATOH, K. and STANDLEY, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, vol. 30, no. 4, pp. 772-780. http://dx.doi.org/10.1093/molbev/mst010. PMid:23329690.
http://dx.doi.org/10.1093/molbev/mst010...
). Aligned sequences were further analyzed with Maximum Parsimony PAUP ver. 4.0b 10 (Felsenstein, 1978FELSENSTEIN, J., 1978. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Biology, vol. 27, no. 4, pp. 401-410. http://dx.doi.org/10.1093/sysbio/27.4.401.
http://dx.doi.org/10.1093/sysbio/27.4.40...
) with 1000 replicate tree bisection-reconnection, branch swapping, and random taxon addition; with MulTrees on and trees saving of 100 as peak value for all replicates. Missing characters were considered as a gap while support was determined using 1000 replicates nonparametric bootstrap method. Program MrBayes 3.2.6 (Ronquist et al., 2012RONQUIST, F., TESLENKO, R., VAN DER MARK, P., AYRES, D.L., DARLING, A., HÖHNA, S., LARGET, B., LIU, L., SUCHARD, M.A. and HUELSENBECK, J.P., 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a largemodel space. Systematic Biology, vol. 61, no. 3, pp. 539-542. http://dx.doi.org/10.1093/sysbio/sys029. PMid:22357727.
http://dx.doi.org/10.1093/sysbio/sys029...
) was employed to perform Bayesian analysis. jModelTest 3.7 (Ebert and Peakall, 2009EBERT, D. and PEAKALL, R., 2009. Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Molecular Ecology Resources, vol. 9, no. 3, pp. 673-690. http://dx.doi.org/10.1111/j.1755-0998.2008.02319.x. PMid:21564725.
http://dx.doi.org/10.1111/j.1755-0998.20...
) was used to select the right model.
3. Result
3.1. Characterization of the cp genomes of G. siniacus and D. velutina
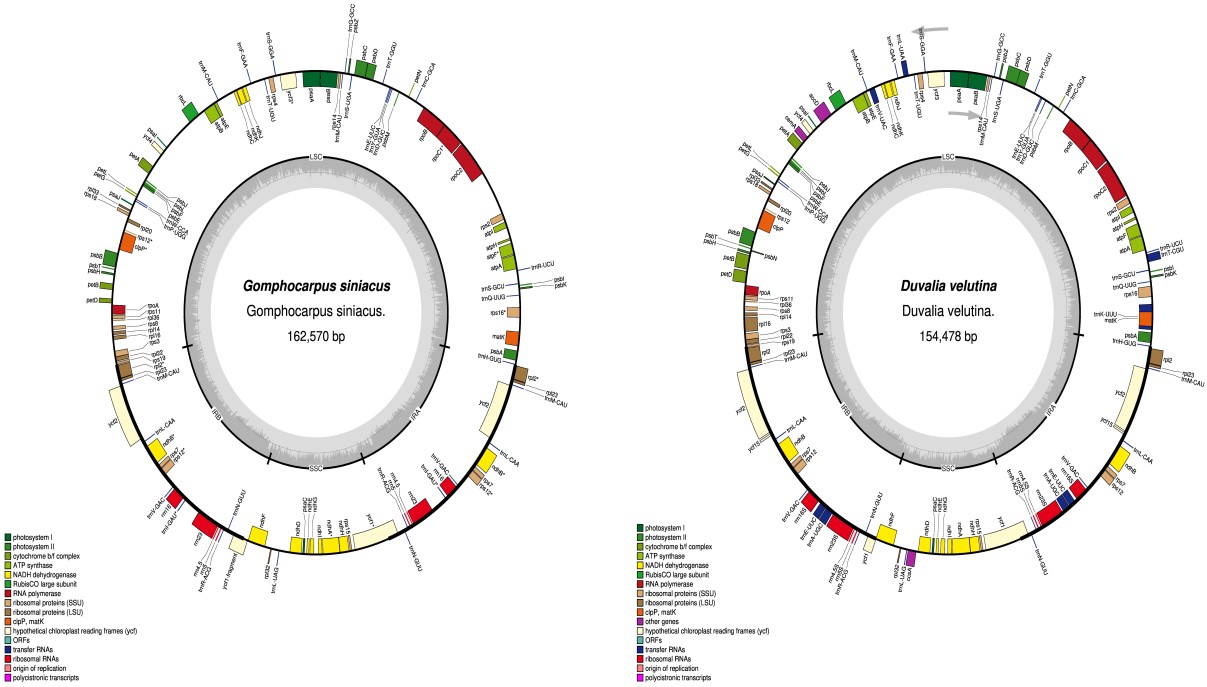
The complete chloroplast genomes of G. siniacus and D. velutina were 154,478 bp and 162,570 bp in size respectively (Table 1 and Figure 1). The two genomes consist of a pair of inverted repeats (25,633 and 26,264 bp); LSC (92,547 and 84,170 bp); SSC (18,757 and 17,780 bp) for D. velutina and G. siniacus respectively (Table 1). Overall GC content of Duvalia velutina is 37.9% while Gomphocarpus siniacus has 38%, while the inverted repeats regions has high GC content ranges from 35.2% to 33.2%. The genome of D. velutina was found to be almost divided into two equal parts between the coding regions and non-coding regions.
The genomes consist of 119 genes (Table 2). The two genomes encode 31tRNA genes, 4 rRNA genes and three pseudo genes (rps19, ycf1 and ycf15). Most of the genes were found to be duplicated at the IR regions of the genomes.
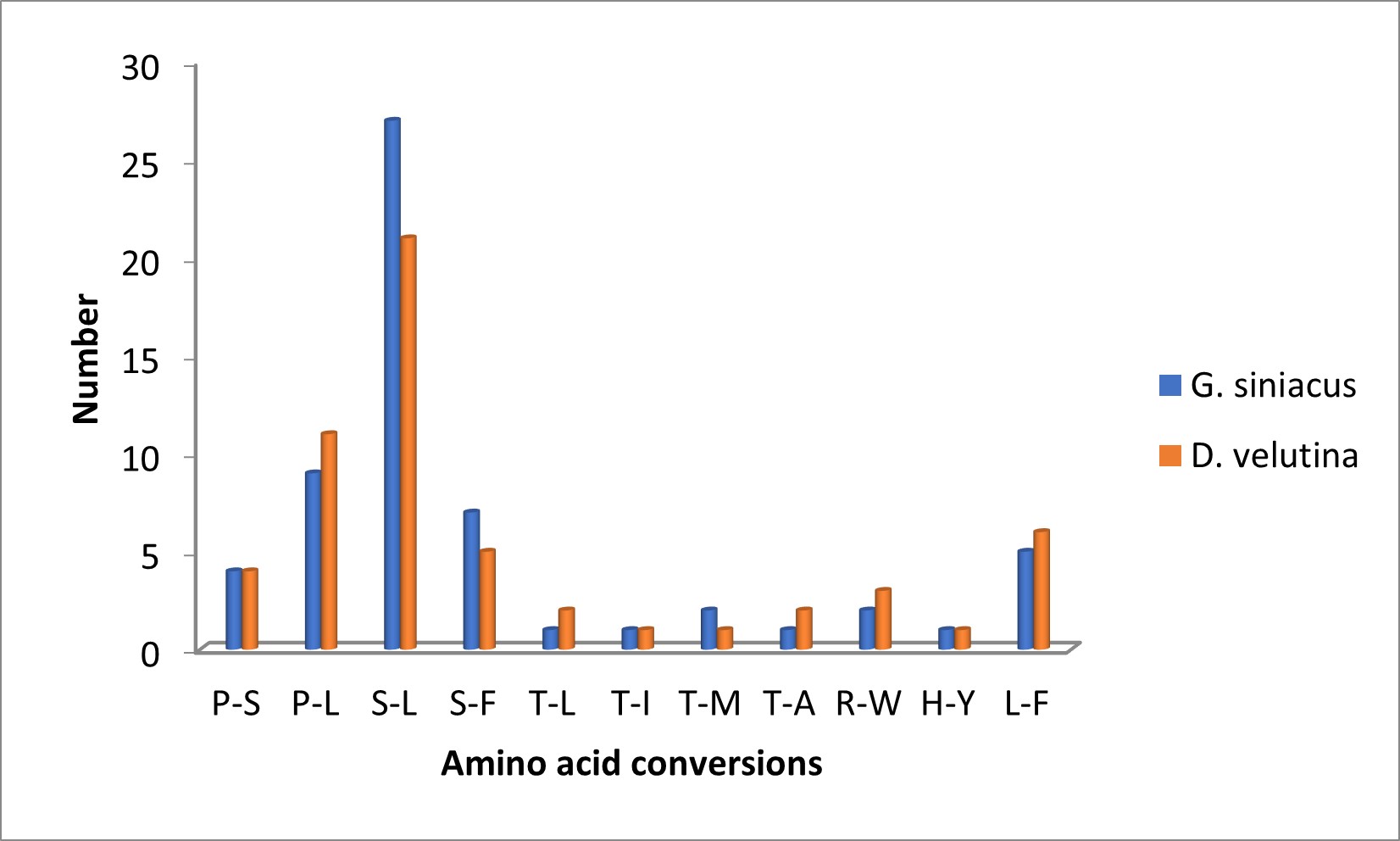
Predictive RNA editing sites indicate high probability in ndhB (8 in D. velutina and 7 in G. siniacus) and rpoB (7 in Both D. velutina and G. siniacus) while matK in D. velutina (3 sites) and ndhA in G. siniacus (5 sites). The conversion rate observed tend to be higher at the initial nucleotides with almost three times than the second nucleotide (Figure 2). Majority of the RNA editing sites were coding for the conversion of serine to Leucine with higher possibility of hydrophobic amino acid valine, phenylalanine, methionine isoleucine and many more. A total of 37 protein coding genes in D. velutina and G. siniacus were predicted for RNA-editing sites 19 were predicted in D. velutina while 21 were predicted in G. siniacus. Total of 8 and 6 genes do not have RNA editing sites in D. velutina and G siniacus chloroplast genomes respectively. The amino acid conversion indicate high Serine to Leucine conversion(S-L)(26), followed by Proline to Leucin (9) and proline to serine (4) and the least conversion were T-L, T-I, T-A and H-Y each with only one potential conversion Figure 2.
3.2. Analysis of cpSSR
Analysis of cpSSR in the sequences of G. siniacus, D. velutina, G. sylvestre and C. procera chloroplast genome (Figure 3 and 4) indicates higher mononucleotide in all the sequences (31 – 99). In G. siniacus tetra-nucleotide are the second highest (13), di-nucleotide (12), tri-nucleotide (10), penta-nucleotide (9) and hexa-nucleotide (1); D. velutina second highest was tri-nucleotide (9), di-nucleotide (5), tetra-nucleotide (3), penta-nucleotide (2) and hexa-nucleotide (1); G. sylvestre di-nucleotide and tetra-nucleotide were second highest with (3 each), tri-nucleotide (2), hexa-nucleotide (1) while no penta-nucleotide was reported; C. procera the second highest was di-nucleotide (16), followed by tetra and hexa-nucleotide (8 each), while the least were tri-nucleotide and tri-nucleotide each with three repeats.
SSR types in G. siniacus, D. velutina, G. sylvestre and C. procera chloroplast genome sequence.
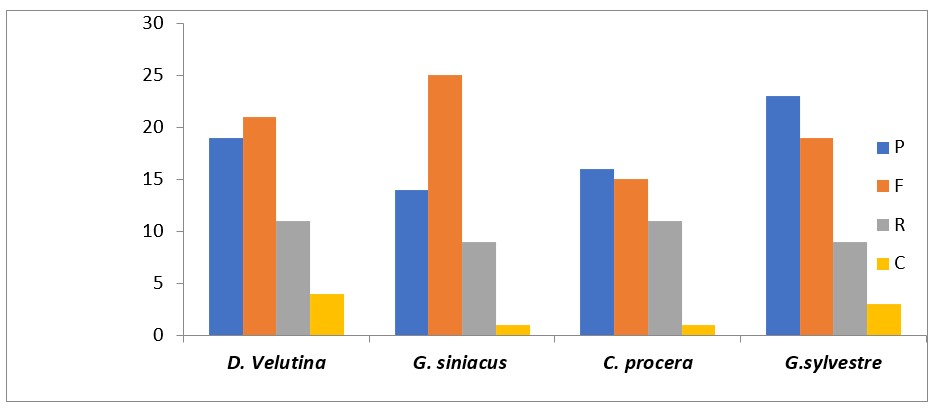
Long Repeats sequences in the four genomes of D. velutina, G. siniacus, C. procera and G. sylvestreFigure 5 generally reveals high number of palindromic and forward repeats. In D. velutina and G. siniacus forward repeats (21 and 25) are higher followed by palindromic (19 and 17), Reverse (11 and 9) and complements (3 and 1); While in C. procera and G. sylvestre forward (15 and 19) repeats are the majority followed by palindromic (16 and 23) followed by Reverse (11 and 9) then complement (1 and 3) being least in the two sequences (Figure 5). AT content of the G. siniacus is (62.1%-63.00%); while in D. velutina it was (61.55%-63.01%); these correspond with the hypothesis that all chloroplast genomes are hardly containing tandem (G) guanine or cytosine (C) but with only polyadenine (polyA) or polythymina (polyT) repeats. The SSRs were also called microsatellites.
Long repeats sequences in D. velutina, G. siniacus, C. procera and G. sylvestre chloroplast genomes. P = palindromic; F = forward; R = reverse; and C = complement. The result of long repeats in Figure 5 indicate highest amount of forward long repeats (21), followed by palindromic (19).
3.3. Substitutions rates analyses
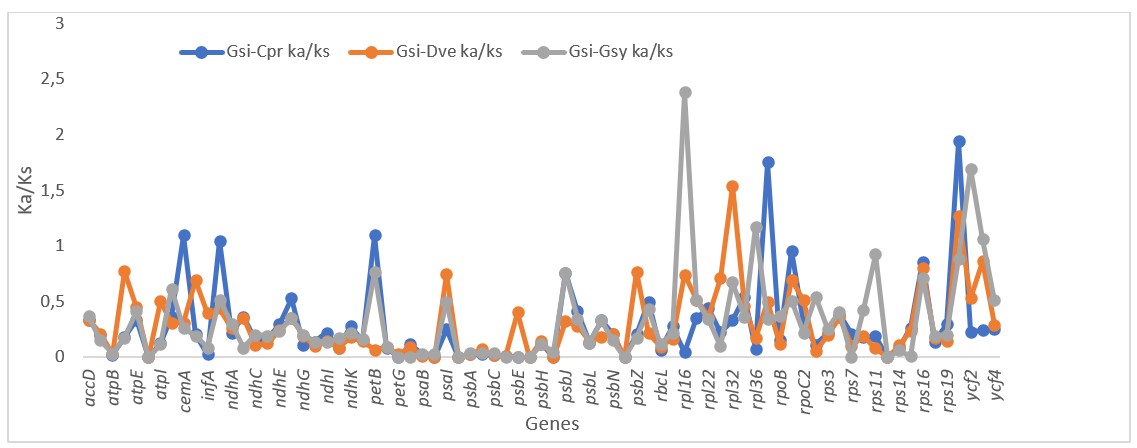
In the study of molecular evolution, the Ka/Ks ratio is used to explain the mechanism of DNA sequence evolution, for the reconstruction of phylogenies, and for the identification of protein-coding genes. It can be used as tool for estimation of the selective pressure of gene evolution, with a Ka/Ks ratio of >1 denoting positive selection and a Ka/Ks ratio of <1 indicating negative selection; a value closer to 1 indicates neutral mutation.Synonymous (Ka) and non-synonymous (Ks) substitution rate and the Ka/Ks ration were determined to evaluate sequence divergence and relative selection in the protein coding genes. The result indicates low sequence divergence in most of the genes (Ks< 0.1) (Figure 6).
3.4. Chloroplast genome comparison of G. siniacus, D. velutina and G. sylvestre and C. procera
The whole chloroplast genomes of G. siniacus, D. velutina and G. sylvestre were compared and annotated C. procera was used as reference sequence (Figure 7) to reveal the features of variations. The output shows there is more variability at the IR regions than the LSC and SSC regions. The coding region is also conserved but non-coding region is less conserved. The most divergent regions are found at the ycf2, psaB, ndhK, ndhB, rpl22, rpoc2, ycf15, petD while at the coding region matK, accD. The Mvista comparison showed that the genomes were conserved with few variations noticed at the non-coding region; the genome of G. siniacus showed good candidacy for the identification and authentication of the taxa on the basis of its structural arrangement. These can be used as molecular markers for the identification of Asclepiadoideae Subfamily and Apocynaceae in general.
Sequence comparisons of four chloroplast genomes using mVISTA programe; with C. procera used as reference genomes.
3.5. Inverted Repeats (IR) junction analysis
Calotropis procera, Gymnema sylvestre, Gomphocarpus siniacus and Duvalia velutina chloroplast genomes border junction comparison (Figure 8) indicates variations between three genes on the basis of their positions; genes such as trnH-GUG, rps19 and ycf1 were observed. trnH-GUG is located at the LSC-IRa border regions of C. procera, Gymnema sylvestre, G. siniacus and D. velutina, genomes while they varied in sizes (3 bp, 1bp, 16bp and 3bp); rps19 and was located at the LSC region in C. procera, G. sylvestre and G. siniacus genomes, while in D. velutina it extended into IRb regions. The disparity due to contractions and expansion of the genome;ycf1 is located at the extensions SSC-IRa border regions in C. procera, G. sylvestre and D. velutina thereby creating pseudo genes between the regions; while in G. siniacus its located at the SSC region.
Comparative chloroplast sequences junctions of LSC, SSC and IR in D. velutina, G. siniacus, and C. procera and G. sylvestre genomes.
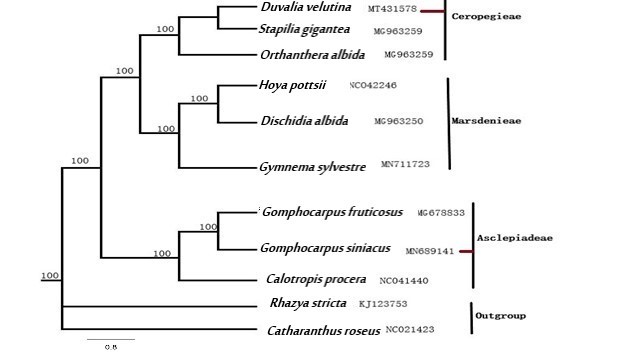
The phylogenetic tree (Figure 9) showed Duvalia and Stapelia are sister taxa and they should be regarded as separate tribes. The sister relationship between Gomphocarpus and Calotropis is also validated. Gomphocarpus and Calotropis were placed in the tribe Asclepiadea as sub tribe.
Phylogenetic tree of 11 taxa based on the complete chloroplast genomes using Bayesian Inference (BI) and Maximum Parsimony (MP) methods; which indicates the relationship within the eleven species of Apocynaceae. The numbers in the branch nodes represent Bootstrap Percentage (BP).
4. Discussion
Two species Asclepiadoideae were assembled and compared with two publicly available species where the sequence alignment, IR contraction and expansion were evaluated. We observed a pseudogenization of ycf1 in G. siniacus sequence. Also the substitution rates were calculated. The four genomes compared were similar in some basic features such as gene contents, number of tRNA and rRNA genes, introns and GC contents. The similarity observed was due to the conserved nature of the chloroplast genome in angiosperm as reported by Ahmed et al. (2013)AHMED, I., MATTHEWS, P.J., BIGGS, P.J., NAEEM, M., MCLENACHAN, P.A. and LOCKHART, P.J., 2013. Identification of chloroplast genome loci suitable for high‐resolution phylogeographic studies of C olocasia esculenta (L.) S chott (A raceae) and closely related taxa. Molecular Ecology Resources, vol. 13, no. 5, pp. 929-937. http://dx.doi.org/10.1111/1755-0998.12128. PMid:23718317.
http://dx.doi.org/10.1111/1755-0998.1212...
, Li et al. (2019)LI, D.-M., ZHAO, C.-Y. and LIU, X.-F., 2019. Complete chloroplast genome sequences of Kaempferia galanga and Kaempferia elegans: molecular structures and comparative analysis. Molecules, vol. 24, no. 3, p. 474. http://dx.doi.org/10.3390/molecules24030474. PMid:30699955.
http://dx.doi.org/10.3390/molecules24030...
, Parks et al. (2009)PARKS, M., CRONN, R. and LISTON, A., 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology, vol. 7, no. 1, p. 84. http://dx.doi.org/10.1186/1741-7007-7-84. PMid:19954512.
http://dx.doi.org/10.1186/1741-7007-7-84...
, Saina et al. (2018)SAINA, J.K., GICHIRA, A.W., LI, Z.-Z., HU, G.-W., WANG, Q.-F. and LIAO, K., 2018. The complete chloroplast genome sequence of Dodonaea viscosa: comparative and phylogenetic analyses. Genetica, vol. 146, no. 1, pp. 101-113. http://dx.doi.org/10.1007/s10709-017-0003-x. PMid:29170851.
http://dx.doi.org/10.1007/s10709-017-000...
. Some species were also varied in their gene number as a result of loss or gain of an intron at either genus level or at family level (Menezes et al., 2018MENEZES, A.P.A., RESENDE-MOREIRA, L.C., BUZATTI, R.S.O., NAZARENO, A.G., CARLSEN, M., LOBO, F.P., KALAPOTHAKIS, E. and LOVATO, M.B., 2018. Chloroplast genomes of Byrsonima species (Malpighiaceae): comparative analysis and screening of high divergence sequences. Scientific Reports, vol. 8, no. 1, p. 2210. http://dx.doi.org/10.1038/s41598-018-20189-4. PMid:29396532.
http://dx.doi.org/10.1038/s41598-018-201...
; Abdullah et al., 2019ABDULLAH, SHAHZADI, I., MEHMOOD, F., ALI, Z., MALIK, M.S., WASEEM, S., MIRZA, B., AHMED, I. and WAHEED, M.T., 2019. Comparative analyses of chloroplast genomes among three Firmiana species: identification of mutational hotspots and phylogenetic relationship with other species of Malvaceae. Plant Gene, vol. 19, p. 100199. http://dx.doi.org/10.1016/j.plgene.2019.100199.
http://dx.doi.org/10.1016/j.plgene.2019....
).
However, positioning of rps19 at the IR region in the G. siniacus (Figure 8) was also observed by Cui et al. (2019CUI, Y., ZHOU, J., CHEN, X., XU, Z., WANG, Y., SUN, W., SONG, J. and YAO, H., 2019. Complete chloroplast genome and comparative analysis of three Lycium (Solanaceae) species with medicinal and edible properties. Gene Reports, vol. 17, p. 100464. http://dx.doi.org/10.1016/j.genrep.2019.100464.
http://dx.doi.org/10.1016/j.genrep.2019....
) and Yaradua et al. (2019YARADUA, S.S., ALZAHRANI, D.A., ALBOKHARY, E.J., ABBA, A. and BELLO, A., 2019. Complete chloroplast genome sequence of Justicia flava: genome comparative analysis and phylogenetic relationships among Acanthaceae. BioMed Research International, vol. 2019, p. 4370258. http://dx.doi.org/10.1155/2019/4370258. PMid:31467890.
http://dx.doi.org/10.1155/2019/4370258...
). Three genes were present in both genomes which is imperative to the findings in other species such as Lycium babarum (Solanaceae) and Swertia mussotii (Gentianaceae) where rps19 andYcf1 were found to be pseudogenes (Cui et al., 2019CUI, Y., ZHOU, J., CHEN, X., XU, Z., WANG, Y., SUN, W., SONG, J. and YAO, H., 2019. Complete chloroplast genome and comparative analysis of three Lycium (Solanaceae) species with medicinal and edible properties. Gene Reports, vol. 17, p. 100464. http://dx.doi.org/10.1016/j.genrep.2019.100464.
http://dx.doi.org/10.1016/j.genrep.2019....
; Xiang et al., 2016XIANG, B., LI, X., QIAN, J., WANG, L., MA, L., TIAN, X. and WANG, Y., 2016. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules, vol. 21, no. 8, p. 1029. http://dx.doi.org/10.3390/molecules21081029. PMid:27517885.
http://dx.doi.org/10.3390/molecules21081...
). This Serine-leucine (S–L) amino acid conversion happens in most angiosperms, as reported in L. barbarum and L. chinense (Cui et al., 2019CUI, Y., ZHOU, J., CHEN, X., XU, Z., WANG, Y., SUN, W., SONG, J. and YAO, H., 2019. Complete chloroplast genome and comparative analysis of three Lycium (Solanaceae) species with medicinal and edible properties. Gene Reports, vol. 17, p. 100464. http://dx.doi.org/10.1016/j.genrep.2019.100464.
http://dx.doi.org/10.1016/j.genrep.2019....
) and also in Dendrobium officinale (Luo et al., 2014) as well as Aristolochia debilis (Zhou et al., 2017ZHOU, J., CHEN, X., CUI, Y., SUN, W., LI, Y., WANG, Y., SONG, J. and YAO, H., 2017. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species. International Journal of Molecular Sciences, vol. 18, no. 9, p. 1839. http://dx.doi.org/10.3390/ijms18091839. PMid:28837061.
http://dx.doi.org/10.3390/ijms18091839...
). . Ycf1 is located at the extensions of IRb and SSC regions there by creating pseudo genes between the regions. IR region is regarded as the most conserved regions in chloroplast genome (Cui et al., 2019CUI, Y., ZHOU, J., CHEN, X., XU, Z., WANG, Y., SUN, W., SONG, J. and YAO, H., 2019. Complete chloroplast genome and comparative analysis of three Lycium (Solanaceae) species with medicinal and edible properties. Gene Reports, vol. 17, p. 100464. http://dx.doi.org/10.1016/j.genrep.2019.100464.
http://dx.doi.org/10.1016/j.genrep.2019....
; Zhou et al., 2017ZHOU, J., CHEN, X., CUI, Y., SUN, W., LI, Y., WANG, Y., SONG, J. and YAO, H., 2017. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species. International Journal of Molecular Sciences, vol. 18, no. 9, p. 1839. http://dx.doi.org/10.3390/ijms18091839. PMid:28837061.
http://dx.doi.org/10.3390/ijms18091839...
; Raubeson and Jansen, 2005RAUBESON, L.A. and JANSEN, R.K., 2005. Chloroplast genomes of plants. In: R.J. HENRY, ed. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Wallingford: CABI Publishing, pp. 45-68. http://dx.doi.org/10.1079/9780851999043.0045.
http://dx.doi.org/10.1079/9780851999043....
).
Overall GC content of the genomes were 37.8% and 40.7% Figure 5 for G. siniacus and D. velutina, also the findings reveals high GC content at the IR region (35.2-33.2%) of the two genomes. This variation was also observed by Cui et al. (2019)CUI, Y., ZHOU, J., CHEN, X., XU, Z., WANG, Y., SUN, W., SONG, J. and YAO, H., 2019. Complete chloroplast genome and comparative analysis of three Lycium (Solanaceae) species with medicinal and edible properties. Gene Reports, vol. 17, p. 100464. http://dx.doi.org/10.1016/j.genrep.2019.100464.
http://dx.doi.org/10.1016/j.genrep.2019....
, Raveendar et al. (2015)RAVEENDAR, S., NA, Y.-W., LEE, J.-R., SHIM, D., MA, K.-H., LEE, S.-Y. and CHUNG, J.-W., 2015. The complete chloroplast genome of Capsicum annuum var. glabriusculum using Illumina sequencing. Molecules, vol. 20, no. 7, pp. 13080-13088. http://dx.doi.org/10.3390/molecules200713080. PMid:26205052.
http://dx.doi.org/10.3390/molecules20071...
, Xiang et al. (2016)XIANG, B., LI, X., QIAN, J., WANG, L., MA, L., TIAN, X. and WANG, Y., 2016. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules, vol. 21, no. 8, p. 1029. http://dx.doi.org/10.3390/molecules21081029. PMid:27517885.
http://dx.doi.org/10.3390/molecules21081...
, in which the authors attributed the variation to the localization of rRNA at IR region. The higher AT content at the third position in the coding was also observed by Cui et al. (2019)CUI, Y., ZHOU, J., CHEN, X., XU, Z., WANG, Y., SUN, W., SONG, J. and YAO, H., 2019. Complete chloroplast genome and comparative analysis of three Lycium (Solanaceae) species with medicinal and edible properties. Gene Reports, vol. 17, p. 100464. http://dx.doi.org/10.1016/j.genrep.2019.100464.
http://dx.doi.org/10.1016/j.genrep.2019....
, Xiang et al. (2016)XIANG, B., LI, X., QIAN, J., WANG, L., MA, L., TIAN, X. and WANG, Y., 2016. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules, vol. 21, no. 8, p. 1029. http://dx.doi.org/10.3390/molecules21081029. PMid:27517885.
http://dx.doi.org/10.3390/molecules21081...
, He et al. (2017)HE, L., QIAN, J., LI, X., SUN, Z., XU, X. and CHEN, S., 2017. Complete chloroplast genome of medicinal plant Lonicera japonica: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules, vol. 22, no. 2, p. 249. http://dx.doi.org/10.3390/molecules22020249. PMid:28178222.
http://dx.doi.org/10.3390/molecules22020...
. This findings was used in the discrimination of chloroplast DNA from the Nuclear and mitochondrial DNA (Shen et al., 2018SHEN, X., GUO, S., YIN, Y., ZHANG, J., YIN, X., LIANG, C., WANG, Z., HUANG, B., LIU, Y., XIAO, S. and ZHU, G., 2018. Complete chloroplast genome sequence and phylogenetic analysis of Aster tataricus. Molecules, vol. 23, no. 10, p. 2426. http://dx.doi.org/10.3390/molecules23102426. PMid:30248930.
http://dx.doi.org/10.3390/molecules23102...
; Clegg et al., 1995CLEGG, M.T., GAUT, B.S., LEARN JUNIOR, G.H. and MORTON, B.R., 1995. Rates and patterns of chloroplast DNA evolution. In: W.M. FITCH and F.J. AYALA, eds. Tempo and mode in evolution: genetics and paleontology 50 years after Simpson. Washington, D.C.: National Academy Press, pp. 215-234.).The GC contents of the cp genomes in this study is similar to the other cp genomes of Apocynaceae and its very much GC-lacking as a result it causes a very much bias towards A/T at the third codon positions (Qian et al., 2013QIAN, J., SONG, J., GAO, H., ZHU, Y., XU, J., PANG, X., YAO, H., SUN, C., LI, X., LI, C., LIU, J., XU, H. and CHEN, S., 2013. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS One, vol. 8, no. 2, p. e57607. http://dx.doi.org/10.1371/journal.pone.0057607. PMid:23460883.
http://dx.doi.org/10.1371/journal.pone.0...
). Mutations occur as a result of translation-preferred codons due to natural selection during evolution of the cp genomes (Yang et al., 2018YANG, Y., ZHU, J., FENG, L., ZHOU, T., BAI, G., YANG, J. and ZHAO, G., 2018. Plastid genome comparative and phylogenetic analyses of the key genera in Fagaceae: highlighting the effect of codon composition bias in phylogenetic inference. Frontiers in Plant Science, vol. 9, p. 82. http://dx.doi.org/10.3389/fpls.2018.00082. PMid:29449857.
http://dx.doi.org/10.3389/fpls.2018.0008...
) RNA editing is an evolutionary process that modify the genetic makeup of a genome by altering the precursor RNA’s nucleotide sequence (Tsudzuki et al., 2001TSUDZUKI, T., WAKASUGI, T. and SUGIURA, M., 2001. Comparative analysis of RNA editing sites in higher plant chloroplasts. Journal of Molecular Evolution, vol. 53, no. 4-5, pp. 327-332. http://dx.doi.org/10.1007/s002390010222. PMid:11675592.
http://dx.doi.org/10.1007/s002390010222...
). This process of the post-transcriptional modification of precursor RNAs to alter their nucleotide sequences (Hoch et al., 1991HOCH, B., MAIER, R.M., APPEL, K., IGLOI, G.L. and KÖSSEL, H., 1991. Editing of a chloroplast mRNA by creation of an initiation codon. Nature, vol. 353, no. 6340, pp. 178-180. http://dx.doi.org/10.1038/353178a0. PMid:1653905.
http://dx.doi.org/10.1038/353178a0...
). It sometimes occurs through the insertion and deletion of nucleotides, or specific nucleotide substitution (mostly C to U conversion) (Hoch et al., 1991HOCH, B., MAIER, R.M., APPEL, K., IGLOI, G.L. and KÖSSEL, H., 1991. Editing of a chloroplast mRNA by creation of an initiation codon. Nature, vol. 353, no. 6340, pp. 178-180. http://dx.doi.org/10.1038/353178a0. PMid:1653905.
http://dx.doi.org/10.1038/353178a0...
). Since the first evidence of RNA editing was found in chloroplast in the rpl2 transcript of maize (Freyer et al., 1997FREYER, R., KIEFER-MEYER, M.-C. and KÖSSEL, H., 1997. Occurrence of plastid RNA editing in all major lineages of land plants. Proceedings of the National Academy of Sciences of the United States of America, vol. 94, no. 12, pp. 6285-6290. http://dx.doi.org/10.1073/pnas.94.12.6285. PMid:9177209.
http://dx.doi.org/10.1073/pnas.94.12.628...
), it has been hunted out and systematically studied in the protein-coding transcripts of majority land plants lineages (Tillich et al., 2005TILLICH, M., FUNK, H.T., SCHMITZ‐LINNEWEBER, C., POLTNIGG, P., SABATER, B., MARTIN, M. and MAIER, R.M., 2005. Editing of plastid RNA in Arabidopsis thaliana ecotypes. The Plant Journal, vol. 43, no. 5, pp. 708-715. http://dx.doi.org/10.1111/j.1365-313X.2005.02484.x. PMid:16115067.
http://dx.doi.org/10.1111/j.1365-313X.20...
), such as Arabidopsis thaliana (Tillich et al., 2010TILLICH, M., BEICK, S. and SCHMITZ-LINNEWEBER, C., 2010. Chloroplast RNA-binding proteins: repair and regulation of chloroplast transcripts. RNA Biology, vol. 7, no. 2, pp. 172-178. http://dx.doi.org/10.4161/rna.7.2.11090. PMid:20215878.
http://dx.doi.org/10.4161/rna.7.2.11090...
), N. tabacum (Yin et al., 2018), Zea mays (Maier et al., 1996MAIER, R.M., ZEITZ, P., KÖSSEL, H., BONNARD, G., GUALBERTO, J.M. and GRIENENBERGER, J.M., 1996. RNA editing in plant mitochondria and chloroplasts. In: W. FILIPOWICZ and T. HOHN, eds. Post-transcriptional control of gene expression in plants. Dordrecht: Springer, pp. 343-365. http://dx.doi.org/10.1007/978-94-009-0353-1_16.
http://dx.doi.org/10.1007/978-94-009-035...
), Oryza sativa (Corneille et al., 2000), D. velutina and P. tomentosa (Abba et al., 2020ABBA, A., ALZAHRANI, D., YARADUA, S. and ALBOKHARI, E.B., 2020. Complete plastome genome of Pergularia tomentosa L. (Asclepiadoideae, Apocynaceae). Mitochondrial DNA. Part B, Resources, vol. 5, no. 1, pp. 566-567. http://dx.doi.org/10.1080/23802359.2019.1710291. PMid:33366649.
http://dx.doi.org/10.1080/23802359.2019....
, 2021ABBA, A., ALZAHRANI, D.A., YARADUA, S.S. and ALBOKHARI, E.J., 2021. Complete chloroplast genome sequencing of Caralluma quadrangula and comparative analysis of the Asclepiadoideae subfamily (Apocynaceae). Journal of Applied Botany and Food Quality, vol. 94, pp. 148-161.).
Most studies noted that start or stop codons were created by RNA editing which result in shortening of the size of translation products (Ozawa et al., 1997OZAWA, S., KOBAYASHI, T., SUGIYAMA, R., HOSHIDA, H., SHIINA, T. and TOYOSHIMA, Y., 1997. Role of PSII-L protein (psbL gene product) on the electron transfer in photosystem II complex. 1. Over-production of wild-type and mutant versions of PSII-L protein and reconstitution into the PSII core complex. Plant Molecular Biology, vol. 34, no. 1, pp. 151-161. http://dx.doi.org/10.1023/A:1005800909495. PMid:9177321.
http://dx.doi.org/10.1023/A:100580090949...
; Wakasugi et al., 1996WAKASUGI, T., HIROSE, T., HORIHATA, M., TSUDZUKI, T., KÖSSEL, H. and SUGIURA, M., 1996. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proceedings of the National Academy of Sciences of the United States of America, vol. 93, no. 16, pp. 8766-8770. http://dx.doi.org/10.1073/pnas.93.16.8766. PMid:8710946.
http://dx.doi.org/10.1073/pnas.93.16.876...
; Yoshinaga et al., 1997YOSHINAGA, K., KAKEHI, T., SHIMA, Y., IINUMA, H., MASUZAWA, T. and UENO, M., 1997. Extensive RNA editing and possible double-stranded structures determining editing sites in the atpB transcripts of hornwort chloroplasts. Nucleic Acids Research, vol. 25, no. 23, pp. 4830-4834. http://dx.doi.org/10.1093/nar/25.23.4830. PMid:9365264.
http://dx.doi.org/10.1093/nar/25.23.4830...
). Also during production of new gene as a result of one striking case (Wakasugi et al., 1996WAKASUGI, T., HIROSE, T., HORIHATA, M., TSUDZUKI, T., KÖSSEL, H. and SUGIURA, M., 1996. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proceedings of the National Academy of Sciences of the United States of America, vol. 93, no. 16, pp. 8766-8770. http://dx.doi.org/10.1073/pnas.93.16.8766. PMid:8710946.
http://dx.doi.org/10.1073/pnas.93.16.876...
); Our findings revealed that there is an codon initiation in psbL gene which is responsible for the production of PSII- L protein (Ozawa et al., 1997OZAWA, S., KOBAYASHI, T., SUGIYAMA, R., HOSHIDA, H., SHIINA, T. and TOYOSHIMA, Y., 1997. Role of PSII-L protein (psbL gene product) on the electron transfer in photosystem II complex. 1. Over-production of wild-type and mutant versions of PSII-L protein and reconstitution into the PSII core complex. Plant Molecular Biology, vol. 34, no. 1, pp. 151-161. http://dx.doi.org/10.1023/A:1005800909495. PMid:9177321.
http://dx.doi.org/10.1023/A:100580090949...
), as previously reported in tobacco (Bock et al., 1993BOCK, R., HAGEMANN, R., KÖSSEL, H. and KUDLA, J., 1993. Tissue-and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids: a new regulatory mechanism? Molecular and General Genetics MGG, vol. 240, no. 2, pp. 238-244. http://dx.doi.org/10.1007/BF00277062. PMid:8355656.
http://dx.doi.org/10.1007/BF00277062...
; Kudla et al., 1992KUDLA, J., IGLOI, G.L., METZLAFF, M., HAGEMANN, R. and KÖSSEL, H., 1992. RNA editing in tobacco chloroplasts leads to the formation of a translatable psbL mRNA by a C to U substitution within the initiation codon. The EMBO Journal, vol. 11, no. 3, pp. 1099-1103. http://dx.doi.org/10.1002/j.1460-2075.1992.tb05149.x. PMid:1547774.
http://dx.doi.org/10.1002/j.1460-2075.19...
) and pepper (Kuntz et al., 1992KUNTZ, M., CAMARA, B., WEIL, J.-H. and SCHANTZ, R., 1992. The psbL gene from bell pepper (Capsicum annuum): plastid RNA editing also occurs in non-photosynthetic chromoplasts. Plant Molecular Biology, vol. 20, no. 6, pp. 1185-1188. http://dx.doi.org/10.1007/BF00028906. PMid:1463853.
http://dx.doi.org/10.1007/BF00028906...
) and spinach (Maier et al., 1996MAIER, R.M., ZEITZ, P., KÖSSEL, H., BONNARD, G., GUALBERTO, J.M. and GRIENENBERGER, J.M., 1996. RNA editing in plant mitochondria and chloroplasts. In: W. FILIPOWICZ and T. HOHN, eds. Post-transcriptional control of gene expression in plants. Dordrecht: Springer, pp. 343-365. http://dx.doi.org/10.1007/978-94-009-0353-1_16.
http://dx.doi.org/10.1007/978-94-009-035...
). RNA editing is common in cp genomes of angiosperms. It usually alters reading frames, mutation, as well as regulation of genes expressions of plants it however, serve as a corrective mechanism in the cp genomes of angiosperms.
RNA editing sites in the sequences of G. siniacus and D. velutina were high from Leucine to serine and mostly the codon conversion from hydrophilic to hydrophobic amino acids were also observed. This has also been reported in other angiosperms by Mehmood et al. (2020)MEHMOOD, F., ABDULLAH, SHAHZADI, I., AHMED, I., WAHEED, M.T. and MIRZA, B., 2020. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics, vol. 112, no. 2, pp. 1522-1530. http://dx.doi.org/10.1016/j.ygeno.2019.08.024. PMid:31470082.
http://dx.doi.org/10.1016/j.ygeno.2019.0...
, Abdullah et al. (2019)ABDULLAH, SHAHZADI, I., MEHMOOD, F., ALI, Z., MALIK, M.S., WASEEM, S., MIRZA, B., AHMED, I. and WAHEED, M.T., 2019. Comparative analyses of chloroplast genomes among three Firmiana species: identification of mutational hotspots and phylogenetic relationship with other species of Malvaceae. Plant Gene, vol. 19, p. 100199. http://dx.doi.org/10.1016/j.plgene.2019.100199.
http://dx.doi.org/10.1016/j.plgene.2019....
. The variation in the sizes of the genomes is as a resultof expansion and contraction of IR borders (Yang et al., 2016YANG, Y., ZHOU, T., DUAN, D., YANG, J., FENG, L. and ZHAO, G., 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Frontiers in Plant Science, vol. 7, p. 959. http://dx.doi.org/10.3389/fpls.2016.00959. PMid:27446185.
http://dx.doi.org/10.3389/fpls.2016.0095...
). This expansion was reported in B. prionitis, in Acanthaceae. The sizes of the inverted repeats were 25,104 bp in D. velutina and 25,461 bp in G. siniacus.
Four sequences were compared for IR borders, where three types of junctions were recognized based on the position of rps19 gene, trnH and ycf1-ndhF positioning. In the first border there is similar orientation of the SSC, LSC and IRa and IRb in D. velutina, C. procera and G. sylvestre. In the G. siniacus sequence there is a clear variation in the orientations of Junctions. Secondly, uniform Border junction was observed in the four sequences while at the third position position a ycf1 in G. siniacus was unique in its position because of its appearance at the forward strand while in D. velutina, C. procera and G. sylvestre it appeared in both forward and reverse strands.
Chloroplast genome has been reported to be much conserved in nature although there is report of variation between species as reported by Yang et al., 2016. The Mvista comparison showed that the genomes were conserved with few variations noticed at the non-coding region; the genome of G. siniacus showed good candidacy for the identification and authentication of the taxa on the basis of its structural arrangement (Rousseau-Gueutin et al., 2015ROUSSEAU-GUEUTIN, M., BELLOT, S., MARTIN, G.E., BOUTTE, J., CHELAIFA, H., LIMA, O., MICHON-COUDOUEL, S., NAQUIN, D., SALMON, A., AINOUCHE, K. and AINOUCHE, M., 2015. The chloroplast genome of the hexaploid Spartina maritima (Poaceae, Chloridoideae): comparative analyses and molecular dating. Molecular Phylogenetics and Evolution, vol. 93, pp. 5-16. http://dx.doi.org/10.1016/j.ympev.2015.06.013. PMid:26182838.
http://dx.doi.org/10.1016/j.ympev.2015.0...
; Yang et al., 2016YANG, Y., ZHOU, T., DUAN, D., YANG, J., FENG, L. and ZHAO, G., 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Frontiers in Plant Science, vol. 7, p. 959. http://dx.doi.org/10.3389/fpls.2016.00959. PMid:27446185.
http://dx.doi.org/10.3389/fpls.2016.0095...
). Alignment of four genomes shows variable regions in the four sequences such as trnH-guG, rbcL, rps16-trnQ and rps19. These can be used as molecular markers for the identification of Asclepiadoideae Subfamily and Apocynaceae in general.
We used the complete cp genomes to reconstitute a Phylogenetic tree and to establish the phylogenetic relationships, as well as tribal positions.The phylogenetic tree showed Duvalia and Stapelia are sister taxa as reported previously (Silva et al., 2012SILVA, U.C.S., RAPINI, A., LIEDE-SCHUMANN, S., RIBEIRO, P.L. and VAN DEN BERG, C., 2012. Taxonomic considerations on Metastelmatinae (Apocynaceae) based on plastid and nuclear DNA. Systematic Botany, vol. 37, no. 3, pp. 795-806. http://dx.doi.org/10.1600/036364412X648733.
http://dx.doi.org/10.1600/036364412X6487...
) therefore should be regarded as separate tribes. The sister relationship between Gomphocarpus and Calotropis is also validated. Gomphocarpus and Calotropis were placed in the tribe Asclepiadeae as sub tribe (Nazar et al., 2019NAZAR, N., CLARKSON, J.J., GOYDER, D., KAKY, E., MAHMOOD, T. and CHASE, M.W., 2019. Phylogenetic relation-ships in Apocynaceae based on nucle-ar PHYA and plastid trnL-F sequences, with a focus on tribal relationships. Caryologia, vol. 72, no. 1, pp. 55-81.; Sinha and Mondal, 2017SINHA, S. and MONDAL, A.K., 2017. A phylogenetic study of floral morphology of some members of Asclepiadaceae R.Br. Annals of Plant Sciences, vol. 6, no. 2, pp. 1546-1559. http://dx.doi.org/10.21746/aps.2017.02.004.
http://dx.doi.org/10.21746/aps.2017.02.0...
). Recently Sinha and Mondal; Nazar et al., 2019 both classified Gomphocarpus under the tribe Asclepiadeae on the basis of molecular trnL-trnF markers. Our results indicate the position of Duvalia as member of the tribe Ceropegieae while Gomphocarpus has been placed under tribe Asclepiadeae.
5. Conclusion
The study involves sequencing and analysis of two species of Asclepiadoideae, G. siniacus and D. velutina (Apocynaceae). The structures of the two genomes were also compared in which different variable regions and SSR markers were unmasked. Also, the gene content arrangements and order were very much conserved. These detailed studies explain the evolutionary relationship among these two genomes which could help in identification, authentication, breeding and evolutionary studies of the family Apocynaceae.
Acknowledgements
The project was financially supported by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, with grant No.D1441-260-130. Therefore, the authors appreciate the gesture.
References
- ABBA, A., ALZAHRANI, D., YARADUA, S. and ALBOKHARI, E.B., 2020. Complete plastome genome of Pergularia tomentosa L. (Asclepiadoideae, Apocynaceae). Mitochondrial DNA. Part B, Resources, vol. 5, no. 1, pp. 566-567. http://dx.doi.org/10.1080/23802359.2019.1710291 PMid:33366649.
» http://dx.doi.org/10.1080/23802359.2019.1710291 - ABBA, A., ALZAHRANI, D.A., YARADUA, S.S. and ALBOKHARI, E.J., 2021. Complete chloroplast genome sequencing of Caralluma quadrangula and comparative analysis of the Asclepiadoideae subfamily (Apocynaceae). Journal of Applied Botany and Food Quality, vol. 94, pp. 148-161.
- ABDULLAH, SHAHZADI, I., MEHMOOD, F., ALI, Z., MALIK, M.S., WASEEM, S., MIRZA, B., AHMED, I. and WAHEED, M.T., 2019. Comparative analyses of chloroplast genomes among three Firmiana species: identification of mutational hotspots and phylogenetic relationship with other species of Malvaceae. Plant Gene, vol. 19, p. 100199. http://dx.doi.org/10.1016/j.plgene.2019.100199
» http://dx.doi.org/10.1016/j.plgene.2019.100199 - AHMED, I., MATTHEWS, P.J., BIGGS, P.J., NAEEM, M., MCLENACHAN, P.A. and LOCKHART, P.J., 2013. Identification of chloroplast genome loci suitable for high‐resolution phylogeographic studies of C olocasia esculenta (L.) S chott (A raceae) and closely related taxa. Molecular Ecology Resources, vol. 13, no. 5, pp. 929-937. http://dx.doi.org/10.1111/1755-0998.12128 PMid:23718317.
» http://dx.doi.org/10.1111/1755-0998.12128 - AMIRYOUSEFI, A., HYVÖNEN, J. and POCZAI, P., 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics, vol. 34, no. 17, pp. 3030-3031. http://dx.doi.org/10.1093/bioinformatics/bty220 PMid:29659705.
» http://dx.doi.org/10.1093/bioinformatics/bty220 - BOCK, R., HAGEMANN, R., KÖSSEL, H. and KUDLA, J., 1993. Tissue-and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids: a new regulatory mechanism? Molecular and General Genetics MGG, vol. 240, no. 2, pp. 238-244. http://dx.doi.org/10.1007/BF00277062 PMid:8355656.
» http://dx.doi.org/10.1007/BF00277062 - BOLGER, A.M., LOHSE, M. and USADEL, B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, vol. 30, no. 15, pp. 2114-2120. http://dx.doi.org/10.1093/bioinformatics/btu170 PMid:24695404.
» http://dx.doi.org/10.1093/bioinformatics/btu170 - BURKILL, H.M., 2004. The useful plants of West tropical Africa Kew: Royal Botanic Gardens, vol. 6.
- CLEGG, M.T., GAUT, B.S., LEARN JUNIOR, G.H. and MORTON, B.R., 1995. Rates and patterns of chloroplast DNA evolution. In: W.M. FITCH and F.J. AYALA, eds. Tempo and mode in evolution: genetics and paleontology 50 years after Simpson Washington, D.C.: National Academy Press, pp. 215-234.
- CORRIVEAU, J.L. and COLEMAN, A.W., 1988. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany, vol. 75, no. 10, pp. 1443-1458. http://dx.doi.org/10.1002/j.1537-2197.1988.tb11219.x
» http://dx.doi.org/10.1002/j.1537-2197.1988.tb11219.x - CUI, Y., ZHOU, J., CHEN, X., XU, Z., WANG, Y., SUN, W., SONG, J. and YAO, H., 2019. Complete chloroplast genome and comparative analysis of three Lycium (Solanaceae) species with medicinal and edible properties. Gene Reports, vol. 17, p. 100464. http://dx.doi.org/10.1016/j.genrep.2019.100464
» http://dx.doi.org/10.1016/j.genrep.2019.100464 - DONG, W., LIU, J., YU, J., WANG, L. and ZHOU, S., 2012. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One, vol. 7, no. 4, p. e35071. http://dx.doi.org/10.1371/journal.pone.0035071 PMid:22511980.
» http://dx.doi.org/10.1371/journal.pone.0035071 - EBERT, D. and PEAKALL, R., 2009. Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Molecular Ecology Resources, vol. 9, no. 3, pp. 673-690. http://dx.doi.org/10.1111/j.1755-0998.2008.02319.x PMid:21564725.
» http://dx.doi.org/10.1111/j.1755-0998.2008.02319.x - FELSENSTEIN, J., 1978. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Biology, vol. 27, no. 4, pp. 401-410. http://dx.doi.org/10.1093/sysbio/27.4.401
» http://dx.doi.org/10.1093/sysbio/27.4.401 - FREYER, R., KIEFER-MEYER, M.-C. and KÖSSEL, H., 1997. Occurrence of plastid RNA editing in all major lineages of land plants. Proceedings of the National Academy of Sciences of the United States of America, vol. 94, no. 12, pp. 6285-6290. http://dx.doi.org/10.1073/pnas.94.12.6285 PMid:9177209.
» http://dx.doi.org/10.1073/pnas.94.12.6285 - HE, L., QIAN, J., LI, X., SUN, Z., XU, X. and CHEN, S., 2017. Complete chloroplast genome of medicinal plant Lonicera japonica: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules, vol. 22, no. 2, p. 249. http://dx.doi.org/10.3390/molecules22020249 PMid:28178222.
» http://dx.doi.org/10.3390/molecules22020249 - HOCH, B., MAIER, R.M., APPEL, K., IGLOI, G.L. and KÖSSEL, H., 1991. Editing of a chloroplast mRNA by creation of an initiation codon. Nature, vol. 353, no. 6340, pp. 178-180. http://dx.doi.org/10.1038/353178a0 PMid:1653905.
» http://dx.doi.org/10.1038/353178a0 - JANSEN, J.J.P., VAN DEN BOSCH, F.A.J. and VOLBERDA, H.W., 2005. Managing potential and realized absorptive capacity: how do organizational antecedents matter? Academy of Management Journal, vol. 48, no. 6, pp. 999-1015. http://dx.doi.org/10.5465/amj.2005.19573106
» http://dx.doi.org/10.5465/amj.2005.19573106 - KATOH, K. and STANDLEY, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, vol. 30, no. 4, pp. 772-780. http://dx.doi.org/10.1093/molbev/mst010 PMid:23329690.
» http://dx.doi.org/10.1093/molbev/mst010 - KATOH, K. and STANDLEY, D.M., 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics, vol. 32, no. 13, pp. 1933-1942. http://dx.doi.org/10.1093/bioinformatics/btw108 PMid:27153688.
» http://dx.doi.org/10.1093/bioinformatics/btw108 - KEARSE, M., MOIR, R., WILSON, A., STONES-HAVAS, S., CHEUNG, M., STURROCK, S., BUXTON, S., COOPER, A., MARKOWITZ, S., DURAN, C., THIERER, T., ASHTON, B., MEINTJES, P. and DRUMMOND, A., 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, vol. 28, no. 12, pp. 1647-1649. http://dx.doi.org/10.1093/bioinformatics/bts199 PMid:22543367.
» http://dx.doi.org/10.1093/bioinformatics/bts199 - KUDLA, J., IGLOI, G.L., METZLAFF, M., HAGEMANN, R. and KÖSSEL, H., 1992. RNA editing in tobacco chloroplasts leads to the formation of a translatable psbL mRNA by a C to U substitution within the initiation codon. The EMBO Journal, vol. 11, no. 3, pp. 1099-1103. http://dx.doi.org/10.1002/j.1460-2075.1992.tb05149.x PMid:1547774.
» http://dx.doi.org/10.1002/j.1460-2075.1992.tb05149.x - KUMAR, S., NEI, M., DUDLEY, J. and TAMURA, K., 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics, vol. 9, no. 4, pp. 299-306. http://dx.doi.org/10.1093/bib/bbn017 PMid:18417537.
» http://dx.doi.org/10.1093/bib/bbn017 - KUNTZ, M., CAMARA, B., WEIL, J.-H. and SCHANTZ, R., 1992. The psbL gene from bell pepper (Capsicum annuum): plastid RNA editing also occurs in non-photosynthetic chromoplasts. Plant Molecular Biology, vol. 20, no. 6, pp. 1185-1188. http://dx.doi.org/10.1007/BF00028906 PMid:1463853.
» http://dx.doi.org/10.1007/BF00028906 - KURTZ, S., CHOUDHURI, J.V., OHLEBUSCH, E., SCHLEIERMACHER, C., STOYE, J. and GIEGERICH, R., 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research, vol. 29, no. 22, pp. 4633-4642. http://dx.doi.org/10.1093/nar/29.22.4633 PMid:11713313.
» http://dx.doi.org/10.1093/nar/29.22.4633 - LANG, Y., WANG, M., XIA, J. and ZHAO, Q., 2018. Effects of soil drought stress on photosynthetic gas exchange traits and chlorophyll fluorescence in Forsythia suspensa. Journal of Forestry Research, vol. 29, no. 1, pp. 45-53. http://dx.doi.org/10.1007/s11676-017-0420-9
» http://dx.doi.org/10.1007/s11676-017-0420-9 - LI, D.-M., ZHAO, C.-Y. and LIU, X.-F., 2019. Complete chloroplast genome sequences of Kaempferia galanga and Kaempferia elegans: molecular structures and comparative analysis. Molecules, vol. 24, no. 3, p. 474. http://dx.doi.org/10.3390/molecules24030474 PMid:30699955.
» http://dx.doi.org/10.3390/molecules24030474 - LI, X., ZHANG, T.-C., QIAO, Q., REN, Z., ZHAO, J., YONEZAWA, T., HASEGAWA, M., CRABBE, M.J.C., LI, J. and ZHONG, Y., 2013. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS One, vol. 8, no. 3, p. e58747. http://dx.doi.org/10.1371/journal.pone.0058747 PMid:23554920.
» http://dx.doi.org/10.1371/journal.pone.0058747 - LIBRADO, P. and ROZAS, J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, vol. 25, no. 11, pp. 1451-1452. http://dx.doi.org/10.1093/bioinformatics/btp187 PMid:19346325.
» http://dx.doi.org/10.1093/bioinformatics/btp187 - LOHSE, M., DRECHSEL, O. and BOCK, R., 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics, vol. 52, no. 5-6, pp. 267-274. http://dx.doi.org/10.1007/s00294-007-0161-y PMid:17957369.
» http://dx.doi.org/10.1007/s00294-007-0161-y - LOWE, T.M. and CHAN, P.P., 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Research, vol. 44, no. W1, pp. W54-W57. http://dx.doi.org/10.1093/nar/gkw413 PMid:27174935.
» http://dx.doi.org/10.1093/nar/gkw413 - MAIER, R.M., ZEITZ, P., KÖSSEL, H., BONNARD, G., GUALBERTO, J.M. and GRIENENBERGER, J.M., 1996. RNA editing in plant mitochondria and chloroplasts. In: W. FILIPOWICZ and T. HOHN, eds. Post-transcriptional control of gene expression in plants Dordrecht: Springer, pp. 343-365. http://dx.doi.org/10.1007/978-94-009-0353-1_16
» http://dx.doi.org/10.1007/978-94-009-0353-1_16 - MASRAHI, Y.S., 2015. A new species of Leptadenia (Apocynaceae) and two other new records from southwestern Saudi Arabia. Saudi Journal of Biological Sciences, vol. 22, no. 5, pp. 631-636. http://dx.doi.org/10.1016/j.sjbs.2015.02.003 PMid:26288569.
» http://dx.doi.org/10.1016/j.sjbs.2015.02.003 - MAYOR, C., BRUDNO, M., SCHWARTZ, J.R., POLIAKOV, A., RUBIN, E.M., FRAZER, K.A., PACHTER, L.S. and DUBCHAK, I., 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics, vol. 16, no. 11, pp. 1046-1047. http://dx.doi.org/10.1093/bioinformatics/16.11.1046 PMid:11159318.
» http://dx.doi.org/10.1093/bioinformatics/16.11.1046 - MEHMOOD, F., ABDULLAH, SHAHZADI, I., AHMED, I., WAHEED, M.T. and MIRZA, B., 2020. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics, vol. 112, no. 2, pp. 1522-1530. http://dx.doi.org/10.1016/j.ygeno.2019.08.024 PMid:31470082.
» http://dx.doi.org/10.1016/j.ygeno.2019.08.024 - MENEZES, A.P.A., RESENDE-MOREIRA, L.C., BUZATTI, R.S.O., NAZARENO, A.G., CARLSEN, M., LOBO, F.P., KALAPOTHAKIS, E. and LOVATO, M.B., 2018. Chloroplast genomes of Byrsonima species (Malpighiaceae): comparative analysis and screening of high divergence sequences. Scientific Reports, vol. 8, no. 1, p. 2210. http://dx.doi.org/10.1038/s41598-018-20189-4 PMid:29396532.
» http://dx.doi.org/10.1038/s41598-018-20189-4 - MOCAN, A., VLASE, L., VODNAR, D.C., BISCHIN, C., HANGANU, D., GHELDIU, A.-M., OPREAN, R., SILAGHI-DUMITRESCU, R. and CRIȘAN, G., 2014. Polyphenolic content, antioxidant and antimicrobial activities of Lycium barbarum L. and Lycium chinense Mill. leaves. Molecules, vol. 19, no. 7, pp. 10056-10073. http://dx.doi.org/10.3390/molecules190710056 PMid:25014533.
» http://dx.doi.org/10.3390/molecules190710056 - MOWER, J.P., 2009. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Research, vol. 37, suppl. 2, pp. W253-W259. http://dx.doi.org/10.1093/nar/gkp337 PMid:19433507.
» http://dx.doi.org/10.1093/nar/gkp337 - NAZAR, N., CLARKSON, J.J., GOYDER, D., KAKY, E., MAHMOOD, T. and CHASE, M.W., 2019. Phylogenetic relation-ships in Apocynaceae based on nucle-ar PHYA and plastid trnL-F sequences, with a focus on tribal relationships. Caryologia, vol. 72, no. 1, pp. 55-81.
- NEUHAUS, H.E. and EMES, M.J., 2000. Nonphotosynthetic metabolism in plastids. Annual Review of Plant Physiology and Plant Molecular Biology, vol. 51, no. 1, pp. 111-140. http://dx.doi.org/10.1146/annurev.arplant.51.1.111 PMid:15012188.
» http://dx.doi.org/10.1146/annurev.arplant.51.1.111 - OZAWA, S., KOBAYASHI, T., SUGIYAMA, R., HOSHIDA, H., SHIINA, T. and TOYOSHIMA, Y., 1997. Role of PSII-L protein (psbL gene product) on the electron transfer in photosystem II complex. 1. Over-production of wild-type and mutant versions of PSII-L protein and reconstitution into the PSII core complex. Plant Molecular Biology, vol. 34, no. 1, pp. 151-161. http://dx.doi.org/10.1023/A:1005800909495 PMid:9177321.
» http://dx.doi.org/10.1023/A:1005800909495 - PARKS, M., CRONN, R. and LISTON, A., 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology, vol. 7, no. 1, p. 84. http://dx.doi.org/10.1186/1741-7007-7-84 PMid:19954512.
» http://dx.doi.org/10.1186/1741-7007-7-84 - QIAN, J., SONG, J., GAO, H., ZHU, Y., XU, J., PANG, X., YAO, H., SUN, C., LI, X., LI, C., LIU, J., XU, H. and CHEN, S., 2013. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS One, vol. 8, no. 2, p. e57607. http://dx.doi.org/10.1371/journal.pone.0057607 PMid:23460883.
» http://dx.doi.org/10.1371/journal.pone.0057607 - QU, X.-J., MOORE, M.J., LI, D.-Z. and YI, T.-S., 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, vol. 15, no. 1, p. 50. http://dx.doi.org/10.1186/s13007-019-0435-7 PMid:31139240.
» http://dx.doi.org/10.1186/s13007-019-0435-7 - RAUBESON, L.A. and JANSEN, R.K., 2005. Chloroplast genomes of plants. In: R.J. HENRY, ed. Plant diversity and evolution: genotypic and phenotypic variation in higher plants Wallingford: CABI Publishing, pp. 45-68. http://dx.doi.org/10.1079/9780851999043.0045
» http://dx.doi.org/10.1079/9780851999043.0045 - RAVEENDAR, S., NA, Y.-W., LEE, J.-R., SHIM, D., MA, K.-H., LEE, S.-Y. and CHUNG, J.-W., 2015. The complete chloroplast genome of Capsicum annuum var. glabriusculum using Illumina sequencing. Molecules, vol. 20, no. 7, pp. 13080-13088. http://dx.doi.org/10.3390/molecules200713080 PMid:26205052.
» http://dx.doi.org/10.3390/molecules200713080 - RODRÍGUEZ-EZPELETA, N., BRINKMANN, H., BUREY, S.C., ROURE, B., BURGER, G., LÖFFELHARDT, W., BOHNERT, H.J., PHILIPPE, H. and LANG, B.F., 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Current Biology, vol. 15, no. 14, pp. 1325-1330. http://dx.doi.org/10.1016/j.cub.2005.06.040 PMid:16051178.
» http://dx.doi.org/10.1016/j.cub.2005.06.040 - RONQUIST, F., TESLENKO, R., VAN DER MARK, P., AYRES, D.L., DARLING, A., HÖHNA, S., LARGET, B., LIU, L., SUCHARD, M.A. and HUELSENBECK, J.P., 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a largemodel space. Systematic Biology, vol. 61, no. 3, pp. 539-542. http://dx.doi.org/10.1093/sysbio/sys029 PMid:22357727.
» http://dx.doi.org/10.1093/sysbio/sys029 - ROUSSEAU-GUEUTIN, M., BELLOT, S., MARTIN, G.E., BOUTTE, J., CHELAIFA, H., LIMA, O., MICHON-COUDOUEL, S., NAQUIN, D., SALMON, A., AINOUCHE, K. and AINOUCHE, M., 2015. The chloroplast genome of the hexaploid Spartina maritima (Poaceae, Chloridoideae): comparative analyses and molecular dating. Molecular Phylogenetics and Evolution, vol. 93, pp. 5-16. http://dx.doi.org/10.1016/j.ympev.2015.06.013 PMid:26182838.
» http://dx.doi.org/10.1016/j.ympev.2015.06.013 - SAINA, J.K., GICHIRA, A.W., LI, Z.-Z., HU, G.-W., WANG, Q.-F. and LIAO, K., 2018. The complete chloroplast genome sequence of Dodonaea viscosa: comparative and phylogenetic analyses. Genetica, vol. 146, no. 1, pp. 101-113. http://dx.doi.org/10.1007/s10709-017-0003-x PMid:29170851.
» http://dx.doi.org/10.1007/s10709-017-0003-x - SHEN, X., GUO, S., YIN, Y., ZHANG, J., YIN, X., LIANG, C., WANG, Z., HUANG, B., LIU, Y., XIAO, S. and ZHU, G., 2018. Complete chloroplast genome sequence and phylogenetic analysis of Aster tataricus. Molecules, vol. 23, no. 10, p. 2426. http://dx.doi.org/10.3390/molecules23102426 PMid:30248930.
» http://dx.doi.org/10.3390/molecules23102426 - SHINOZAKI, K., OHME, M., TANAKA, M., WAKASUGI, T., HAYASHIDA, N., MATSUBAYASHI, T., ZAITA, N., CHUNWONGSE, J., OBOKATA, J., YAMAGUCHI‐SHINOZAKI, K., OHTO, C., TORAZAWA, K., MENG, B.Y., SUGITA, M., DENO, H., KAMOGASHIRA, T., YAMADA, K., KUSUDA, J., TAKAIWA, F., KATO, A., TOHDOH, N., SHIMADA, H. and SUGIURA, M., 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. The EMBO Journal, vol. 5, no. 9, pp. 2043-2049. http://dx.doi.org/10.1002/j.1460-2075.1986.tb04464.x PMid:16453699.
» http://dx.doi.org/10.1002/j.1460-2075.1986.tb04464.x - SILVA, U.C.S., RAPINI, A., LIEDE-SCHUMANN, S., RIBEIRO, P.L. and VAN DEN BERG, C., 2012. Taxonomic considerations on Metastelmatinae (Apocynaceae) based on plastid and nuclear DNA. Systematic Botany, vol. 37, no. 3, pp. 795-806. http://dx.doi.org/10.1600/036364412X648733
» http://dx.doi.org/10.1600/036364412X648733 - SINHA, S. and MONDAL, A.K., 2017. A phylogenetic study of floral morphology of some members of Asclepiadaceae R.Br. Annals of Plant Sciences, vol. 6, no. 2, pp. 1546-1559. http://dx.doi.org/10.21746/aps.2017.02.004
» http://dx.doi.org/10.21746/aps.2017.02.004 - THIEL, T., MICHALEK, W., VARSHNEY, R. and GRANER, A., 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theoretical and Applied Genetics, vol. 106, no. 3, pp. 411-422. http://dx.doi.org/10.1007/s00122-002-1031-0 PMid:12589540.
» http://dx.doi.org/10.1007/s00122-002-1031-0 - TILLICH, M., BEICK, S. and SCHMITZ-LINNEWEBER, C., 2010. Chloroplast RNA-binding proteins: repair and regulation of chloroplast transcripts. RNA Biology, vol. 7, no. 2, pp. 172-178. http://dx.doi.org/10.4161/rna.7.2.11090 PMid:20215878.
» http://dx.doi.org/10.4161/rna.7.2.11090 - TILLICH, M., FUNK, H.T., SCHMITZ‐LINNEWEBER, C., POLTNIGG, P., SABATER, B., MARTIN, M. and MAIER, R.M., 2005. Editing of plastid RNA in Arabidopsis thaliana ecotypes. The Plant Journal, vol. 43, no. 5, pp. 708-715. http://dx.doi.org/10.1111/j.1365-313X.2005.02484.x PMid:16115067.
» http://dx.doi.org/10.1111/j.1365-313X.2005.02484.x - TILLICH, M., LEHWARK, P., PELLIZZER, T., ULBRICHT-JONES, E.S., FISCHER, A., BOCK, R. and GREINER, S., 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Research, vol. 45, no. W1, pp. W6-W11. http://dx.doi.org/10.1093/nar/gkx391 PMid:28486635.
» http://dx.doi.org/10.1093/nar/gkx391 - TONTI‐FILIPPINI, J., NEVILL, P.G., DIXON, K. and SMALL, I., 2017. What can we do with 1000 plastid genomes? The Plant Journal, vol. 90, no. 4, pp. 808-818. http://dx.doi.org/10.1111/tpj.13491 PMid:28112435.
» http://dx.doi.org/10.1111/tpj.13491 - TSUDZUKI, T., WAKASUGI, T. and SUGIURA, M., 2001. Comparative analysis of RNA editing sites in higher plant chloroplasts. Journal of Molecular Evolution, vol. 53, no. 4-5, pp. 327-332. http://dx.doi.org/10.1007/s002390010222 PMid:11675592.
» http://dx.doi.org/10.1007/s002390010222 - WAKASUGI, T., HIROSE, T., HORIHATA, M., TSUDZUKI, T., KÖSSEL, H. and SUGIURA, M., 1996. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proceedings of the National Academy of Sciences of the United States of America, vol. 93, no. 16, pp. 8766-8770. http://dx.doi.org/10.1073/pnas.93.16.8766 PMid:8710946.
» http://dx.doi.org/10.1073/pnas.93.16.8766 - WICKE, S., SCHNEEWEISS, G.M., DEPAMPHILIS, C.W., MÜLLER, K.F. and QUANDT, D., 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Molecular Biology, vol. 76, no. 3-5, pp. 273-297. http://dx.doi.org/10.1007/s11103-011-9762-4 PMid:21424877.
» http://dx.doi.org/10.1007/s11103-011-9762-4 - XIANG, B., LI, X., QIAN, J., WANG, L., MA, L., TIAN, X. and WANG, Y., 2016. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules, vol. 21, no. 8, p. 1029. http://dx.doi.org/10.3390/molecules21081029 PMid:27517885.
» http://dx.doi.org/10.3390/molecules21081029 - XIE, Y., WU, G., TANG, J., LUO, R., PATTERSON, J., LIU, S., HUANG, W., HE, G., GU, S., LI, S., ZHOU, X., LAM, T.-W., LI, Y., XU, X., WONG, G.K.-S. and WANG, J., 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics, vol. 30, no. 12, pp. 1660-1666. http://dx.doi.org/10.1093/bioinformatics/btu077 PMid:24532719.
» http://dx.doi.org/10.1093/bioinformatics/btu077 - YANG, Y., ZHOU, T., DUAN, D., YANG, J., FENG, L. and ZHAO, G., 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Frontiers in Plant Science, vol. 7, p. 959. http://dx.doi.org/10.3389/fpls.2016.00959 PMid:27446185.
» http://dx.doi.org/10.3389/fpls.2016.00959 - YANG, Y., ZHU, J., FENG, L., ZHOU, T., BAI, G., YANG, J. and ZHAO, G., 2018. Plastid genome comparative and phylogenetic analyses of the key genera in Fagaceae: highlighting the effect of codon composition bias in phylogenetic inference. Frontiers in Plant Science, vol. 9, p. 82. http://dx.doi.org/10.3389/fpls.2018.00082 PMid:29449857.
» http://dx.doi.org/10.3389/fpls.2018.00082 - YARADUA, S.S., ALZAHRANI, D.A., ALBOKHARY, E.J., ABBA, A. and BELLO, A., 2019. Complete chloroplast genome sequence of Justicia flava: genome comparative analysis and phylogenetic relationships among Acanthaceae. BioMed Research International, vol. 2019, p. 4370258. http://dx.doi.org/10.1155/2019/4370258 PMid:31467890.
» http://dx.doi.org/10.1155/2019/4370258 - YOSHINAGA, K., KAKEHI, T., SHIMA, Y., IINUMA, H., MASUZAWA, T. and UENO, M., 1997. Extensive RNA editing and possible double-stranded structures determining editing sites in the atpB transcripts of hornwort chloroplasts. Nucleic Acids Research, vol. 25, no. 23, pp. 4830-4834. http://dx.doi.org/10.1093/nar/25.23.4830 PMid:9365264.
» http://dx.doi.org/10.1093/nar/25.23.4830 - ZHOU, J., CHEN, X., CUI, Y., SUN, W., LI, Y., WANG, Y., SONG, J. and YAO, H., 2017. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species. International Journal of Molecular Sciences, vol. 18, no. 9, p. 1839. http://dx.doi.org/10.3390/ijms18091839 PMid:28837061.
» http://dx.doi.org/10.3390/ijms18091839
Publication Dates
-
Publication in this collection
29 Aug 2022 -
Date of issue
2024
History
-
Received
09 Oct 2021 -
Accepted
20 July 2022