Abstracts
Coffee is one of the most valuable agricultural commodities and ranks second on international trade exchanges. The genus Coffea belongs to the Rubiaceae family which includes other important plants. The genus contains about 100 species but commercial production is based only on two species, Coffea arabica and Coffea canephora that represent about 70 % and 30 % of the total coffee market, respectively. The Brazilian Coffee Genome Project was designed with the objective of making modern genomics resources available to the coffee scientific community, working on different aspects of the coffee production chain. We have single-pass sequenced a total of 214,964 randomly picked clones from 37 cDNA libraries of C. arabica, C. canephora and C. racemosa, representing specific stages of cells and plant development that after trimming resulted in 130,792, 12,381 and 10,566 sequences for each species, respectively. The ESTs clustered into 17,982 clusters and 32,155 singletons. Blast analysis of these sequences revealed that 22 % had no significant matches to sequences in the National Center for Biotechnology Information database (of known or unknown function). The generated coffee EST database resulted in the identification of close to 33,000 different unigenes. Annotated sequencing results have been stored in an online database at <A HREF="http://www.lge.ibi.unicamp.br/cafe">http://www.lge.ibi.unicamp.br/cafe</A>. Resources developed in this project provide genetic and genomic tools that may hold the key to the sustainability, competitiveness and future viability of the coffee industry in local and international markets.
Coffea; cDNA; EST; transcriptome
O café é um dos principais produtos agrícolas, sendo considerado o segundo item em importância do comércio internacional de "commodities". O gênero Coffea pertence à família Rubiaceae que também inclui outras plantas importantes. Este gênero contém aproximadamente 100 espécies, mas a produção comercial é baseada somente em duas espécies, Coffea arabica e Coffea canephora, que representam aproximadamente 70 % e 30 % do mercado total de café, respectivamente. O Projeto Genoma Café Brasileiro foi desenvolvido com o objetivo de disponibilizar os modernos recursos da genômica à comunidade científica e aos diferentes segmentos da cadeia produtiva do café. Para isso, foram seqüenciados 214.964 clones escolhidos aleatoriamente de 37 bibliotecas de cDNA de C. arabica, C. canephora e C. racemosa representando estádios específicos do desenvolvimento de células e de tecidos do cafeeiro, resultando em 130.792, 12.381 e 10.566 seqüências de cada espécie, respectivamente, após processo de trimagem. Os ESTs foram agrupados em 17.982 contigs e em 32.155 singletons. A comparação destas seqüências pelo programa BLAST revelou que 22 % não tiveram nenhuma similaridade significativa às seqüências no banco de dados do National Center for Biotechnology Information (de função conhecida ou desconhecida). A base de dados de ESTs do cafeeiro resultou na identificação de cerca de 33.000 unigenes diferentes. Os resultados de anotação das seqüências foram armazenados em base de dados "online" em <A HREF="http://www.lge.ibi.unicamp.br/cafe">http://www.lge.ibi.unicamp.br/cafe</A>. Os recursos desenvolvidos por este projeto disponibilizam ferramentas genéticas e genômicas que podem ser decisivas para a sustentabilidade, a competitividade e a futura viabilidade da agroindústria cafeeira nos mercados interno e externo.
Coffea; cDNA; EST; transcritoma
MINIREVIEW
Brazilian coffee genome project: an EST-based genomic resource
Projeto Genoma Brasileiro Café: recursos genômicos baseados em ESTs
Luiz Gonzaga Esteves VieiraI,* * These authors contributed equally to this work. § Corresponding author: lvieira@iapar.br Current Address: 1 Universidade Estadual de Ponta Grossa, Campus Uvaranas, 84030-900, Ponta Grossa, PR, Brazil; 2 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Quitandinha, 25651-075, Petrópolis, RJ, Brazil; 3 Alellyx Applied Genomics, Rod. Anhanguera, Km 104, 13067-850, Campinas, SP, Brazil; ,§ * These authors contributed equally to this work. § Corresponding author: lvieira@iapar.br Current Address: 1 Universidade Estadual de Ponta Grossa, Campus Uvaranas, 84030-900, Ponta Grossa, PR, Brazil; 2 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Quitandinha, 25651-075, Petrópolis, RJ, Brazil; 3 Alellyx Applied Genomics, Rod. Anhanguera, Km 104, 13067-850, Campinas, SP, Brazil; ; Alan Carvalho AndradeII,* * These authors contributed equally to this work. § Corresponding author: lvieira@iapar.br Current Address: 1 Universidade Estadual de Ponta Grossa, Campus Uvaranas, 84030-900, Ponta Grossa, PR, Brazil; 2 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Quitandinha, 25651-075, Petrópolis, RJ, Brazil; 3 Alellyx Applied Genomics, Rod. Anhanguera, Km 104, 13067-850, Campinas, SP, Brazil; ; Carlos Augusto ColomboIII,* * These authors contributed equally to this work. § Corresponding author: lvieira@iapar.br Current Address: 1 Universidade Estadual de Ponta Grossa, Campus Uvaranas, 84030-900, Ponta Grossa, PR, Brazil; 2 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Quitandinha, 25651-075, Petrópolis, RJ, Brazil; 3 Alellyx Applied Genomics, Rod. Anhanguera, Km 104, 13067-850, Campinas, SP, Brazil; ; Ana Heloneida de Araújo MoraesII; Ângela MethaII; Angélica Carvalho de OliveiraII; Carlos Alberto LabateIV; Celso Luis MarinoVIII; Claúdia de Barros Monteiro-VitorelloVI,2 * These authors contributed equally to this work. § Corresponding author: lvieira@iapar.br Current Address: 1 Universidade Estadual de Ponta Grossa, Campus Uvaranas, 84030-900, Ponta Grossa, PR, Brazil; 2 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Quitandinha, 25651-075, Petrópolis, RJ, Brazil; 3 Alellyx Applied Genomics, Rod. Anhanguera, Km 104, 13067-850, Campinas, SP, Brazil; ; Damares de Castro MonteII; Éder GigliotiIX; Edna Teruko KimuraX; Eduardo RomanoII; Eiko Eurya KuramaeXI; Eliana Gertrudes Macedo LemosXII; Elionor Rita Pereira de AlmeidaII; Érika C. JorgeV; Érika V. S. AlbuquerqueII; Felipe Rodrigues da SilvaII; Felipe VineckyII; Haiko Enok SawazakiIII; Hamza Fahmi A. DorryXIV; Helaine CarrerVII; Ilka Nacif AbreuXV; João A. N. BatistaII; João Batista TeixeiraII; João Paulo KitajimaXVII; Karem Guimarães XavierIV; Liziane Maria de LimaII; Luis Eduardo Aranha de CamargoVI; Luiz Filipe Protasio PereiraXVIII; Luiz Lehmann CoutinhoV; Manoel Victor Franco LemosXIII; Marcelo Ribeiro RomanoIV,1 * These authors contributed equally to this work. § Corresponding author: lvieira@iapar.br Current Address: 1 Universidade Estadual de Ponta Grossa, Campus Uvaranas, 84030-900, Ponta Grossa, PR, Brazil; 2 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Quitandinha, 25651-075, Petrópolis, RJ, Brazil; 3 Alellyx Applied Genomics, Rod. Anhanguera, Km 104, 13067-850, Campinas, SP, Brazil; ; Marcos Antonio MachadoXIX; Marcos Mota do Carmo CostaII; Maria Fátima Grossi de SáII; Maria Helena S. GoldmanXX; Maria Inês T. FerroXII; Maria Laine Penha TinocoII; Mariana C. OliveiraXXI; Marie-Anne Van SluysXXI; Milton Massao ShimizuXV; Mirian Perez MalufXXII; Mirian Therezinha Souza da EiraXXIII; Oliveiro Guerreiro FilhoIII; Paulo ArrudaXXIV; Paulo MazzaferaXV; Pilar Drummond Sampaio Correa MarianiXVI; Regina L.B.C. de OliveiraXXV; Ricardo HarakavaXXVI; Silvia Filippi BalbaoXV; Siu Mui TsaiXXVII; Sonia Marli Zingaretti di MauroXII; Suzana Neiva SantosII; Walter José SiqueiraIII; Gustavo Gilson Lacerda CostaXXVIII; Eduardo Fernandes FormighieriXXVIII; Marcelo Falsarella CarazzolleXXVIII,* * These authors contributed equally to this work. § Corresponding author: lvieira@iapar.br Current Address: 1 Universidade Estadual de Ponta Grossa, Campus Uvaranas, 84030-900, Ponta Grossa, PR, Brazil; 2 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Quitandinha, 25651-075, Petrópolis, RJ, Brazil; 3 Alellyx Applied Genomics, Rod. Anhanguera, Km 104, 13067-850, Campinas, SP, Brazil; ; Gonçalo Amarante Guimarães PereiraXXVIII,* * These authors contributed equally to this work. § Corresponding author: lvieira@iapar.br Current Address: 1 Universidade Estadual de Ponta Grossa, Campus Uvaranas, 84030-900, Ponta Grossa, PR, Brazil; 2 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Quitandinha, 25651-075, Petrópolis, RJ, Brazil; 3 Alellyx Applied Genomics, Rod. Anhanguera, Km 104, 13067-850, Campinas, SP, Brazil;
ILaboratório de Biotecnologia Vegetal (LBI), IAPAR, CP 481, 86001-970, Londrina, PR, Brazil
IINúcleo de Biotecnologia-NTBio, Embrapa Recursos Genéticos e Biotecnologia, Parque Estação Biológica, CP 02372, 70770-900, Brasília, DF, Brazil
IIIInstituto Agronômico de Campinas, CP 28, 13001-970, Campinas, SP, Brazil
IVDepartamento de Genética, Escola Superior de Agricultura Luiz de Queiroz, USP, USP, 13418-900, Piracicaba, SP, Brasil
VDepartamento de Zootecnia, Escola Superior de Agricultura Luiz de Queiroz, USP, USP, 13418-900, Piracicaba, SP, Brasil
VIDepartamento de Entomologia, Fitopatologia e Zoologia Agrícola, Escola Superior de Agricultura Luiz de Queiroz, USP, USP, 13418-900, Piracicaba, SP, Brasil
VIIDepartamento de Ciências Biológicas , Escola Superior de Agricultura Luiz de Queiroz, USP, USP, 13418-900, Piracicaba, SP, Brasil
VIIIDepartamento de Genética, Instituto de Biociências,UNESP, 18618000, Botucatu SP, Brazil
IXCentro de Ciências Agrárias, Universidade Federal de São Carlos, 13600-970, Araras, SP, Brazil
XInstituto de Ciências Biomédicas, USP, 05508-000, São Paulo, SP, Brazil
XIDepartamento de Defesa Fitossanitária, Faculdade de Ciências Agronômicas, UNESP, CP 237, 18603-970, Botucatu SP, Brazil
XIIDepartamento de Tecnologia, Faculdade de Ciências Agrárias e Veterinárias de Jaboticabal, UNESP, 14884-900, Jaboticabal, SP, Brazil
XIIIDepartamento de Biologia Aplicada à Agropecuária, Faculdade de Ciências Agrárias e Veterinárias de Jaboticabal, UNESP, 14884-900, Jaboticabal, SP, Brazil
XIVDepartamento de Bioquímica, Instituto de Química, USP, 05513-970, São Paulo, SP, Brazil
XVDepartamento de Fisiologia Vegetal, Instituto de Biologia, UNICAMP, CP 6109, 13083-970, Campinas, SP, Brasil
XVIFaculdade de Engenharia Química, Instituto da Computação, UNICAMP, CP 6109, 13083-970, Campinas, SP, Brasil
XVIILaboratório de Bioinformática, Instituto da Computação, UNICAMP, CP 6109, 13083-970, Campinas, SP, Brasil
XVIIIEmbrapa Café, IAPAR, CP 481, 86001-970, Londrina, PR, Brazil
XIXCentro APTA de Citros Sylvio Moreira, IAC, CP 04, 13490-970, Cordeirópolis SP, Brazil
XXDepartamento de Biologia, Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto, USP, 14040-901, Ribeirão Preto, SP, Brazil
XXIDepartamento de Botânica, Instituto de Biociências, Universidade de São Paulo, 05508-900, São Paulo, SP, Brazil
XXIIEmbrapa Café, Instituto Agronômico de Campinas, CP 28, 13001-970, Campinas, SP, Brazil
XXIIIEmbrapa Café, Núcleo de Biotecnologia-NTBio, Embrapa Recursos Genéticos e Biotecnologia, Parque Estação Biológica, CP 02372, 70770-900, Brasília, Brazil
XXIVCentro de Biologia Molecular e Engenharia Genética, UNICAMP, CP 6010, 13083-970, Campinas, SP, Brazil
XXVNúcleo Integrado de Biotecnologia, Universidade de Mogi das Cruzes, 08780-911, Mogi das Cruzes, SP, Brazil
XXVICentro de Sanidade Vegetal, Instituto Biológico de São Paulo, 04014-002, São Paulo, SP, Brazil
XXVIICentro de Energia Nuclear na Agricultura, USP, CP 96, 13400-970, Piracicaba, SP, Brazil
XXVIIILaboratório de Genômica e Expressão, Instituto de Biologia, UNICAMP, 13083-970, Campinas, SP, Brazil
ABSTRACT
Coffee is one of the most valuable agricultural commodities and ranks second on international trade exchanges. The genus Coffea belongs to the Rubiaceae family which includes other important plants. The genus contains about 100 species but commercial production is based only on two species, Coffea arabica and Coffea canephora that represent about 70 % and 30 % of the total coffee market, respectively. The Brazilian Coffee Genome Project was designed with the objective of making modern genomics resources available to the coffee scientific community, working on different aspects of the coffee production chain. We have single-pass sequenced a total of 214,964 randomly picked clones from 37 cDNA libraries of C. arabica, C. canephora and C. racemosa, representing specific stages of cells and plant development that after trimming resulted in 130,792, 12,381 and 10,566 sequences for each species, respectively. The ESTs clustered into 17,982 clusters and 32,155 singletons. Blast analysis of these sequences revealed that 22 % had no significant matches to sequences in the National Center for Biotechnology Information database (of known or unknown function). The generated coffee EST database resulted in the identification of close to 33,000 different unigenes. Annotated sequencing results have been stored in an online database at http://www.lge.ibi.unicamp.br/cafe. Resources developed in this project provide genetic and genomic tools that may hold the key to the sustainability, competitiveness and future viability of the coffee industry in local and international markets.
Key words:Coffea, cDNA, EST, transcriptome.
RESUMO
O café é um dos principais produtos agrícolas, sendo considerado o segundo item em importância do comércio internacional de "commodities". O gênero Coffea pertence à família Rubiaceae que também inclui outras plantas importantes. Este gênero contém aproximadamente 100 espécies, mas a produção comercial é baseada somente em duas espécies, Coffea arabica e Coffea canephora, que representam aproximadamente 70 % e 30 % do mercado total de café, respectivamente. O Projeto Genoma Café Brasileiro foi desenvolvido com o objetivo de disponibilizar os modernos recursos da genômica à comunidade científica e aos diferentes segmentos da cadeia produtiva do café. Para isso, foram seqüenciados 214.964 clones escolhidos aleatoriamente de 37 bibliotecas de cDNA de C. arabica, C. canephora e C. racemosa representando estádios específicos do desenvolvimento de células e de tecidos do cafeeiro, resultando em 130.792, 12.381 e 10.566 seqüências de cada espécie, respectivamente, após processo de trimagem. Os ESTs foram agrupados em 17.982 contigs e em 32.155 singletons. A comparação destas seqüências pelo programa BLAST revelou que 22 % não tiveram nenhuma similaridade significativa às seqüências no banco de dados do National Center for Biotechnology Information (de função conhecida ou desconhecida). A base de dados de ESTs do cafeeiro resultou na identificação de cerca de 33.000 unigenes diferentes. Os resultados de anotação das seqüências foram armazenados em base de dados "online" em http://www.lge.ibi.unicamp.br/cafe. Os recursos desenvolvidos por este projeto disponibilizam ferramentas genéticas e genômicas que podem ser decisivas para a sustentabilidade, a competitividade e a futura viabilidade da agroindústria cafeeira nos mercados interno e externo.
Palavras-chave:Coffea, cDNA, EST, transcritoma.
INTRODUCTION
Coffee is an important agricultural commodity produced in more than 60 countries. It generates a turnover of US$10-12 billion per year and ranks second on international trade exchanges, representing a significant source of income to several developing countries in Africa, Asia and Latin America. Brazil, Vietnam and Colombia are responsible for about 50 % of the world-coffee production, and Brazil alone responds for more than one third of the global coffee production and exports. This fact ranks coffee amongst the most important commodities in the Brazilian trade balance.
The genus Coffea belongs to the Rubiaceae family, found throughout the tropics, which includes other important plants. About 100 species of the genus Coffea have been identified so far (Bridson and Verdcourt, 1988), most of them trees and shrubs growing at low altitudes in the tropical rain forests of Africa and Asia (Sondahl et al., 1992). Commercially, production relies only on two species, Coffea arabica L and Coffea canephora Pierre ex Froehner, which represent about 70 % and 30 % of the total coffee market, respectively. All known species are diploid (2n=22 chromosomes) and obligate outbreeders with self-incompatibility systems, except for C. arabica which is allotetraploid (2n=4x=44) and self-fertile at approximately 90 %. (Charrier and Berthaud, 1985).
C. arabica, providing Arabica coffee, was first described by Linnaeus in 1753.The botanical evidence indicates that the coffee plant C. arabica originated on the plateaus of central Ethiopia where it still grows wild. The species C. arabica L. is endemic in Southwest Ethiopia and probably originates from a relatively recent cross between Coffea eugenoides and Coffea canephora (Lashermes et al., 1993) as indicated by chloroplast restriction fragment length polymorphism (RFLP) analyses (Lashermes et al. 1996). The nuclear DNA content of C. arabica determined by flow cytometry is 2.4 pg, or 2X=1158 Mb (Arumuganathan et al., 1991). C. arabica is the most cultivated species, occupying 75 % of the coffee plantations around the world. The quality of the beverage is potentially excellent, being known in the trade as mild coffee. Several cultivars have been described for C. arabica, but because of the narrow genetic basis of the species, phenotype differences among the cultivars are due mainly to single gene mutations.
The species C. canephora is the diploid species most widely cultivated around the world. It is self-sterile and cross-pollinated and consequently displays much more variability than C. arabica. C. canephora is better adapted to warm and humid equatorial climates and is frequently cultivated in low to medium altitudes. Robusta coffee is grown in West and Central Africa, throughout Southeast Asia and in Brazil, where it is also known as Conilon. The quality of the beverage made from C. canephora is generally regarded as inferior to that made of C. arabica. However, C. canephora is more resistant to adverse conditions than Arabica, particularly to several diseases and pests. Another diploid coffee species originating from Mozambique, C. racemosa, is characterized as having low caffeine content, high drought tolerance and resistance to leaf-miner (Clarke and Macrae, 1988), and has been used in breeding programs for introgression of important agronomic traits to C. arabica (Guerreiro et al., 1991).
The cultivation of the Arabica coffee began about five hundred years ago in Yemen and reached the southeast of Asia approximately in 1700. In the beginning of the 18th century, progenies of a single plant were taken from Indonesia to Europe and later to America (Chevalier and Dagron, 1928; Carvalho, 1945). Originating from other introductions that took place from Yemen to Brazil, seeds of two different cultivars, Typica and Bourbon, constitute the main genetic basis of all cultivated coffee planted in Brazil and other countries (Krug et al., 1939; Carvalho et al., 1993).
Coffee has long been bred with the view of improving important agronomic characteristics such as flowering, yield, bean size, cup quality, caffeine content and disease and drought resistance. Despite solid efforts, the progress in coffee breeding using conventional approaches has been slow due to many factors such as the narrow genetic basis of cultivated coffee, the lack of genetic markers and efficient screening tools, as well as the long time taken for generation advancement.
The recent development of applied technologies in biology is leading to an enormous production of information in the area of plant genomics, through the sequencing of different organisms. Large-scale sequencing of cDNAs to produce Expressed Sequence Tags (ESTs) and comparing the resulting sequences with public databases has become the method of choice for the rapid and cost-effective generation of data on the coding capacity of genomes and for the potential identification of new genes. For the same reason, several sequencing projects of plant species, such as the Sugar Cane EST Genome Project (SUCEST) accomplished by the ONSA group (Organization for Nucleotide Sequencing and Analysis) (Arruda, 2001) have been carried out in Brazil.
Coffea genomes are large in comparison with the current plant models, Arabidopsis and rice. While the coffee genome may probably have similarities to gene motifs already identified in small-genome plants, the larger genome size of coffee makes it unlikely to anticipate a complete genome-sequencing effort of any species of the genus Coffea in the near future, despite the recent increases in DNA-sequencing capacity of modern equipment. Therefore, large-scale discovery, isolation and analysis of gene function in coffee and its relatives must rely on other, less direct methods. The partial sequencing of anonymous cDNA clones (Expressed sequence tags - ESTs) is a rapid and cost-effective method for generating data on the coding capacity of genomes and, for this reason, has become the fastest growing segment of the public DNA databases (Wolfsberg and Landsman, 1997). In plants, the EST approach was initially used for the model species A. thaliana (Höfte et al., 1993) and rice (Yamamoto and Sasaki, 1997). Subsequently, a large variety of EST sequences from other species have been deposited in the dbEST (http://www.ncbi.nlm.nih.gov/dbEST/). Recently, an EST database based on sequences from approximately 47,000 cDNA clones and with a special focus on developing seeds of C. canephora has also been released (Chenwei et al., 2005).
The Brazilian Coffee Genome Project was designed to develop and deploy useful tools for gene discovery and functional genetic analysis in coffee and related species and to aid in the advance of knowledge on the structure and evolution of the coffee genome. The generated coffee EST database from C. arabica, C. canephora and C. racemosa resulted in the identification of more than 30 thousand different unigenes and will facilitate genetic studies on coffee. This basic information provides a very valuable resource for studies on the biology and physiology of coffee plants that will considerably enhance the isolation and characterization of important agronomic genes for genetic improvement of Coffea.
Project organization and goals
The Brazilian Coffee Genome Project was formulated in 2002 through a cooperative agreement signed between the Brazilian Coffee Research and Development Consortium (CBP&D-Café), a national consortium of 40 public Universities and Research Institutes, the Brazilian Enterprise for Agricultural Research (Embrapa), the São Paulo State Research Support Foundation (FAPESP) and the Permanent Forum for University-Company Relations (UNIEMP). The CBP&D was responsible for the central coordination of the project, but all three institutions supporting this initiative also appointed one project coordinator each with managerial responsibilities in the project aimed at facilitating the maintenance of the information flow from the network of laboratories involved in the Coffee Genome Project.
The initial goal of this project was the development of a large database of ESTs, with a minimum of 200,000 reads and a Unigene set composed of 25,000 genes. Assuming that the number of gene motifs is similar among the angiosperm genomes, this number would theoretically represents about 2/3 of all the gene motifs present in the coffee genomes.
The infrastructure assembled for sequencing was already established by the AEG (Agriculture and Environment Genomes), a network of several laboratories located at different research institutions in São Paulo state, and funded by both FAPESP and Embrapa Recursos Genéticos e Biotecnologia, Brasília. Laboratories from these two groups were also responsible for supervising and coordinating all aspects of cDNA library construction, such as cDNA size selection, cloning, clone-picking and clone library storage, sequencing and sequence submission to the bioinformatics center. Each group was assigned the cloning and sequencing of 100,000 reads.
The Laboratório de Genômica e Expressão (LGE) of the State University at Campinas (http://www.lge.ibi.unicamp.br/) was designated as the central bioinformatics facility to house the Coffee Genome sequence database and coordinate all aspects related to sequence submission, performance and productivity of the sequencing groups, data storage, BLAST analysis and clustering. For safety reasons, a replica of the raw data was transferred to the Embrapa Recursos Genéticos e Biotecnologia's bioinformatics group.
Total cost of the project and the committed institutional efforts was shared among the CBP&D-Café, EMBRAPA and FAPESP in the proportions of 50 %, 25 % and 25 %, respectively. The access of the database is free for six public universities and research institutes linked to FAPESP and for organizations and research institutes that are members of the CBP&D-Café, which in turn grant the opportunity for free access to more than 700 scientists from 40 institutions that develop coffee research in Brazil through collaborative partnerships. Data access restrictions are applied to any other user of the EST database, subject to the approval of the Coffee Genome Project Directive Committee. For this purpose, specific contractual conditions have been established regarding intellectual-propriety rights derived from having access to this information.
cDNA libraries and sequencing
The Instituto Agronômico de Campinas (IAC), which possesses a significant germplasm collection of Coffea species, supplied the material for the construction of cDNA libraries covering a wide range of tissues, developmental stages, and plant material submitted to biotic and abiotic stress conditions. The cDNA libraries constructed by the AEG group used plant material from C. arabica cv. Mundo Novo and cv. Catuaí, while those constructed at Embrapa Recursos Genéticos e Biotecnologia were made from tissues and organs from C. arabica cv. Catuaí (table 1). Also, EST libraries were made from tissues of C. canephora and C. racemosa lines belonging to the Instituto Capixaba de Pesquisa, Assistência Técnica e Extensão Rural (INCAPER) and IAC's collection, respectively.
Total RNA was extracted from coffee tissues at different developmental stages and also submitted to different stress conditions. Poly(A)+ RNA was purified from total RNA using the Oligotex Kit (Quiagen), following the manufacturers directions. The mRNA purity and integrity were estimated by absorbance at 260/280 nm and agarose gel electrophoresis. cDNA libraries were constructed using the SuperScript Plasmid System and Plasmid Cloning Kit (Invitrogen) with about 1-2 µg poly(A)+ RNA. The efficiency of cDNA synthesis was monitored with radioactive nucleotides. cDNA were size fractionated on a Sepharose CL-2B column. Aliquots of each fraction were eletrophoresed in agarose gel to determine the size range of cDNAs. Fractions containing cDNA larger than 500 pb were ligated into pSPORT1 and pSPORT6 vectors (Invitrogen) at the SalI-NotI site. The resulting plasmids were transformed in E. coli DH10B or DH5a cells (Invitrogen) by electroporation.
Plasmid DNA was purified using a modified alkaline lysis method (Sambrook et al., 1989). Sequencing reactions were conducted using the ABI BigDye Terminator Sequencing kit (Applied Biosystems). cDNA inserts were sequenced from the 5' end with T7 promoter primer (5'-TAATACGACTCACTATAGGG-3') or M13 Rev in the pSPORT1 vector or with SP6 primer (5'- ATTTAGGTGACACTATAG-3') in the pSPORT6. Sequencing reaction products were analyzed on ABI 3700 sequencers (Applied Biosystems).
Picking of the clones and storage of stocks were carried out at the Brazilian Clone Collection Center (BCCC) in the case of libraries constructed by the AEG group. For the construction of all libraries, no procedure to eliminate differences in transcript representation was adopted.
Sequencing of coffee EST libraries was carried out by 25 laboratories located at Research Institutes and Universities belonging to the AEG system and at Embrapa Recursos Genéticos e Biotecnologia with 96-lane sequencers (ABI 3700), using standard protocols. Raw sequences and base confidence scores were obtained from chromatogram files using the program Phred (Ewing and Green, 1998; Ewing et al., 1998). Sequences accepted in the project had more than 250 bases with Phred quality > 20. The number of sequences collected for each library was determined by monitoring the redundancy level of produced sequences.
Bioinformatics and database construction
In general, bioinformatics of EST projects includes such services as the organization, storage, integration, and analysis of biological information. The objectives of the Laboratório de Genômica e Expressão (LGE) bioinformatics group were (a) to provide appropriate database methods for the data generated for the sequencing groups in São Paulo and Brasília; (b) to provide adequate security measures to ensure the integrity of the data; (c) to organize and present the data in such a way that authorized users can readily extract meaningful information from it and (d) to develop user-friendly interfaces to access the core data.
The first bioinformatics objective of the Brazilian Coffee Genome Project was to establish the means by which the various forms of the core data could be stored. The diverse sources of the sequences submitted required personnel that were knowledgeable in the nature of the data, as well as in the collection, manipulation, presentation and sharing of the bioinformatics data. There was also a need for security of the core data, which requires restricted access and backup, and there was a need for users to be able to access the data on demand.
The second bioinformatics objective was related to the handling of core data to meet the needs of users. For this, a relational database was developed in order to record and readily access the core information. To keep costs to a minimum in a centralized databasing location while making the database amenable to a broad group of users, a MySQL database (http://www.mysql.com/) was preferred for the relational database. EST assembly and the viewing of assemblies, as well as consensus sequences were the prime goals in bioinformatics of the coffee genome project.
Delivery of the information produced by the Brazilian Coffee Genome Project can be retrieved via the internet at the project site (http://www.lge.ibi.unicamp.br/cafe/). The advantage of web-based delivery is that anyone with an internet connection can have access, but this advantage is counterbalanced by the risk of crashes of a single centralized facility and sometimes slow speed of information access. For these reasons, besides LGE, all the core sequence data is also maintained at Embrapa Recursos Genéticos e Biotecnologia bioinformatics group (http://www.cenargen.embrapa.br/biotec/genomacafe/). Also, the possibility of handling the data by two different bioinformatics groups allows the development of derivatives of the core data (e.g., gene specific oligonucleotides, protein sequences, promoters, gene expression data etc) to meet the most frequent needs of users through databases that may be customized to take into consideration the preferences of coffee investigators.
Database analysis
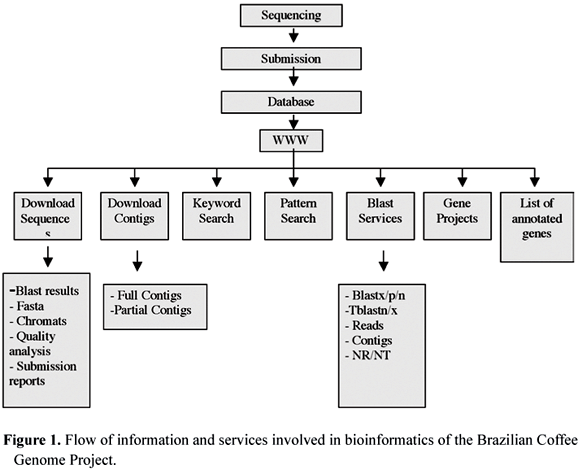
For functional annotation of ESTs and categorization of contigs, the masked (http://www.phrap.com/) and trimmed sequences (Telles and da Silva, 2001) were compared with the protein sequences stored at NCBI databases (National Center for Biotechnology Information), particularly to the NR-(Non-redundant) database (http://www.ncbi.nlm.nih.gov/blast/html/blastcgihelp.shtml#databases). Also, several off the shelf databasing and extraction tools were developed by the LGE team to fulfill many of the initial needs in coffee genomics. The procedures for submitting, processing, storing and analyzing the data are summarized in figure 2.
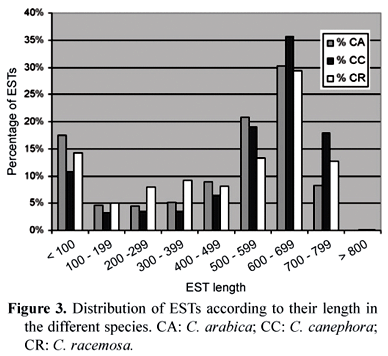
An overall sequencing efficiency of 70 % was obtained, including failures due to false-positive vector-only clones, short-insert clones, low-intensity or no-labeling reads, low-quality reads etc. The final results of the sequencing of EST libraries done by 26 laboratories produced a total of 214,964 reads, distributed among the three Coffea species selected in the project. The quality of the submitted sequences is an important piece of information to validate a database. The majority of the ESTs analyzed at end of the sequencing stage of the Coffee Genome Project had lengths above 500 bp with Phred quality >= 20 (figure 3).
As for any EST project, unwanted sequences are produced such as ribosomal sequences, poly-A fragments, low quality and short sequences, and slippage that all needed to be remove to avoid the introduction of irrelevant information into the EST database. The trimming was carried out with reads from C. arabica, C. canephora and C. racemosa that resulted in 130,792, 12,381 and 10,566 sequences (respectively), with the number of removed sequences summarized in table 2 according to each class.
Clustering and assembly of these ESTs using the CAP3 program (Huang and Madan, 1999) was done separated by species, resulting in 14,886 clusters and 24,426 singletons from C. arabica, 2,147clusters and 4,622 singletons for C. canephora, and 949 clusters and 3,107 singletons for C. racemosa . Close to sixty percent of the 17,982 contigs and singletons presented a size length between 700 and 900 bp (figure 4). Due to the short-length attributes of part of the ESTs that had been produced, some singlets may have failed to merge into contigs and, therefore, the total number of "unigenes" might be overestimated. Of the contigs in C. arabica, the majority (86.7 %) was represented by two to ten ESTs. Due to the small number of clones for C. canephora and C. racemosa that were produced, the percentage of contigs with a higher number of ESTs in that range was superior to C. arabica (97.8 % and 97.5 %, respectively) (figure 5).
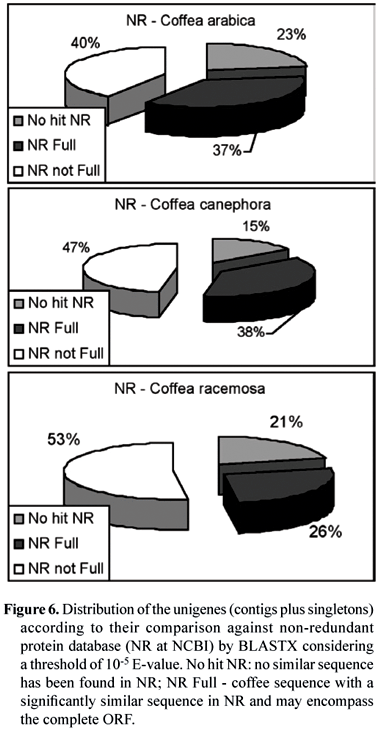
Regarding the cDNA libraries from C. arabica, C. canephora and C. racemosa, the sequences were analyzed for their similarity with known genes by BLASTX (Altschul et al, 1990) against NR (figure 6), considering 10-5 for the E-value as threshold for identity. It is interesting to note that there is a very similar partition between known and unknown sequences (No hit NR) for the three species. Moreover, among the sequences that generated hits, a significant number comprehends cDNA with the complete ORF inside (full length NR Full).
Close to 11,000 contigs from C. arabica (29 % of the dataset) lacked significant similarity to any sequence based on the three ontological principles of Molecular Function, Biological Process and Cellular Component and broad categories developed for plant gene annotations by the Gene Ontology (GO) Consortium (ftp://ftp.geneontology.org/go/GO_slims/). C. canephora and C. racemosa had the same percentage of hits by GO (figure 7).
EST-based functional analysis
Coffee breeding, which is carried out through the traditional methods of hybridization and selection of superior progenies, has achieved relative success in satisfying the needs of the coffee industry. Certainly, the value of conventional breeding should not be overlooked, but linked efforts of both molecular techniques and traditional breeding can offer alternatives for making selective breeding more predictable and precise, reducing the time for obtaining new genotypes. Nowadays, the comprehensive examination of an organism that is afforded by functional genomics has changed the way one identifies genes and proteins with potential roles in a particular biological process without any a priori knowledge of their function. As with conventional breeding, the main objective is to describe and exploit the genetic diversity that is present in coffee species.
The access to coffee gene sequence information brings new perspectives and approaches to carry out biological research. Genome related databases, as the one made available by the Brazilian Coffee Genome Project, have become an invaluable asset for the scientific community to move onto the use of a number of these new technologies. Ultimately, through the use of the coffee EST dataset, genetic markers can be found for breeding programs, coffee genes can be cataloged in association with their location on the genome, the study of gene function and how activity of the gene products fits into complex metabolic pathways can be facilitated, and the regulation of the genes in response to different developmental and environmental stimuli can be examined holistically.
The evaluation of certain characteristics of interest in coffee requires a great deal of time because it can only be carried out on coffee trees after 10-15 years. Particularly, in C. arabica, the limited genetic diversity present among elite cultivars planted all over the world is the consequence of few introductions (Pearl et al., 2004). This lack of genetic variability in the gene pool of Arabica coffee limits the potential for germplasm improvement of this species. Therefore, finding new traits that add value to agricultural crops and their products has immense value in the agribusiness.
Due to the knowledge gathered on the coffee genome, these problems can be alleviated by searching for genetic markers closely linked to the candidate genes expressing these characteristics. The detection of such markers permits the screening of large numbers of coffee trees for a gene of interest when the plants are still at early stages of development and may reduce the number of backcrosses required to obtain quality traits (Lashermes et al., 1997). Also, marker-assisted selection for important but complex traits, which are often difficult to select in routine breeding programs, will enhance coffee breeding programs in terms of better-focused problems and save time and resources.
Molecular markers allow for the extension of conventional breeding methods with one important difference, that is the transfer of genetic information in a more precise and controlled manner. In coffee, molecular marker technology has already been implemented in germplasm characterization and management (Sera et al., 2003; Aga et al., 2005; Prakash et al., 2005; Maluf et al., 2005), detecting gene introgression in breeding populations (Prakash et al., 2002; Prakash et al., 2004; Herrera et al., 2004), describing coffee phylogeny with related species (Lashermes et al., 1999;Anthony et al., 2002) and in marker assisted breeding (Bertrand et al., 2001).
Among the molecular markers currently available, the SSRs (Simple Sequence Repeats), or microsatellite, have been extensively used due to their resolution and polymorphism levels. These characteristics make these molecular markers efficient tools for the genetic mapping, linkage studies, genotype identification and conservation of germplasm, pedigree analyses, marker assisted selection, and analysis of DNA libraries for gene cloning (Rufino et al., 2005). In coffee, the SSRs markers are not broadly used due to the limited numbers of primers presently available for this plant. The availability of massive amounts of coffee nucleotide sequence data will certainly offer an alternative to identify microsatellite motifs, which would be much more expensive through conventional laboratory protocols. In the coffee EST data set, a number of SSRs are present in transcripts that can now be readily mapped using existing breeding populations, and such studies are currently underway (Colombo and Caixeta, personal communication). Furthermore, specific genes of interest can be studied for variation within coffee species, allowing their assignment to coffee linkage maps.
Coffee physical maps bridge gaps between genetic maps and gene location. In this way, the availability of coffee BAC libraries will make possible the alignment of physical and genetic maps, bringing along continuity from phenotype to genotype (Noir et al., 2004; Leroy et al., 2005). Furthermore, the combination of EST database and BAC libraries may help to isolate genes through positional cloning.
One of the major objectives of the Brazilian Coffee Genome Project was to provide a tool for creation of transcriptional profiles as they appear in different tissues and as they change in response to development (Gaspari-Pezzopane et al., 2005; Geromel et al., 2005), biotic (Brandalise et al., 2005) and abiotic stresses (Vinecky et al., 2005). To help accomplish these studies, it is necessary to have powerful technologies available that allow the analysis of mRNA transcription patterns of thousands of genes in a single experiment (Kuhn, 2001).
Gene arrays (Lockhart and Winzeler, 2000) hybridized with mRNA populations from a variety of coffee tissues, organs and developmental stages may provide a genome-wide database of the transcriptional changes during plant growth that ultimately determine resistance to pests and diseases, productivity, and quality attributes of the coffee trees and fruits. Using this screening method, solutions for specific agronomic constraints may be found not only through new cultivar development but also by changes in crop management, harvesting, and post-harvest practices. For the construction of arrays, a set of UNIGENE sequences has to be available for use in the analysis of temporal or spatial expression profiles. Recent work to devise a minimal clone set that represents all transcripts found in the Brazilian Coffee Genome Project was carried out by Sales et al. (2005). In this effort, a single relational database containing close to 33,000 putative transcripts was organized, allowing its use in diverse platforms and languages.
Proteomics as used to identify proteins in complex mixtures is only effective when a sequenced and annotated genome is available or Unigene sets become established (Rounsley et al., 1996). Proteomics is complementary to the ESTs because it also focuses on gene products. Proteomic studies consist of profiling the protein expression levels found in samples derived from different cultivars, tissue types, cultivation or post-harvest conditions in order to understand which proteins may be responsible for a trait of commercial significance, such as pathogens (Andrade et al., 2005), stress tolerance (Vincent et al., 2005) and food quality (Hajduch et al., 2005).
Proteomic characterizations of the coffee genotypes may also be used to validate results derived from DNA arrays and EST studies by verifying protein expression and thereby permit the subsequent coordination of gene transcription with protein expression. Such results can be used to establish baseline protein expression levels, and to identify constitutively expressed proteins that will be used as standards for comparing results derived from different cultivars or crop management conditions.
One of the effective ways to carry out studies on gene function at the morphological, biochemical and physiological level is to establish regulated expression systems of native genes in plants. The cloning of coffee regulatory sequences opens up the possibility of understanding the molecular mechanisms that regulate cellular/developmental processes and production of coffee metabolites at the biochemical and molecular levels, and provides the possibility of using regulatory elements to manipulate expression of entire metabolic pathways.
At the moment, only a few regulatory sequences for some coffee genes (Aldwinckle and Gaitan, 2002, 2004; Marraccini et al., 1999, 2003; Satyanarayana et al., 2005) have been identified. One of the most effective ways to obtain clones for promoter analysis of genes is from large insert genomic libraries. The construction of BAC libraries (Noir et al., 2004; Leroy et al., 2005) in addition to the already available EST sequences may greatly speed up the process of identification and isolation of important genetic control elements in coffee (promoters, enhancers, silencers etc). A highly efficient transformation system in coffee is an important complementary technology for evaluating promoter function.
Production of genetically modified plants is one of the techniques that opens new perspectives to coffee improvement, allowing the fast incorporation of desirable characteristics into elite cultivars. Despite the fact that the discussions on plant transformation are mainly centered on the commercial applications, for the scientific community, transgenic plants are important tools to study various aspects of plant sciences (Pereira, 2000). The enormous amounts of DNA sequence information available in the coffee EST data set opens up new experimental opportunities for functional genomic analysis.
Although genetic transformation procedures for coffee have been established (Hatanaka et al., 1999; Leroy et al., 2000; Ribas et al., 2005), the current technology has serious limitations, including low efficiency and throughput, which is still a key limitation for the widespread use of this technology (see: Genetic Transformation of Coffee, Ribas et al., in this issue). Successful genetic transformation of coffee is still limited to characters controlled by major genes and to transgenic plants that have been produced for insect resistance (Leroy et al., 2000), low caffeine content in seeds (Ogita et al., 2003) and herbicide resistance (Ribas et al., submitted). Based on the current public understanding of this technology, characteristics with low variability in the Coffea gene pool or of great appeal to consumers, such as delayed fruit ripening, resistance to pests and diseases (e.g., coffee borer, nematodes, coffee berry disease, leaf rust, Xylella), tolerance to abiotic stress and enhanced health benefits such as disease-fighting compounds, are the main candidates for academic work in future years.
CONCLUSION
The Brazilian Coffee Genome Project briefly presented here provides the genomic tools required for applied research to address the various constraints associated with the economic production of the coffee industry, mainly regarding the development of new cultivars. When combined with progress made in the development of in vitro technologies required for genetic transformation, data made available by the Coffee Genome Project may place the development of coffee cultivars on the future research agenda through the use of these new genetic technologies.
It is our belief that the Coffee EST database will not be limited to cultivar development applications, but will make a decisive contribution to other applied supplementary applications such as transcriptional profiling and proteomic analyses, leading to a better understanding of the way plants cope with biotic and abiotic stresses. Practical problems faced by the coffee agribusiness, represented by farmers, roasters, processors, exporters and specialty coffee associations, such as control of pre- and post-harvest physiological factors involved in quality, disease and pests control, management of plant response to water deficit, and elevated production costs can be partially overcome by integrated efforts of genomics research and breeding. Also, improvements through coffee genomic research may result in increased consumption and better health value of the beverage through new value-added products derived from coffee (e.g., nutraceutics, oils and flavors).
Finally, with the use of the coffee EST set it may be predicted that the integration of gene discovery, marker development and gene deployment become routine practices in Brazilian coffee research programs. Currently, genome annotation is being carried out by different institutions of the CBP&D-Café to improve the information in the database of the Coffee Genome Project. Annotating EST records will allow the coffee scientific community to use EST databases as an opportunity for gene discovery. Further efforts by the Coffee Genome Project bioinformatics groups may include assembly of ESTs to form Unigene sequences, complete gene sequences, gene specific oligonucleotides, alignment of gene sequences with related genes from other organisms, grouping of genes according to expression pattern and function, genetic linkage maps and physical maps.
Acknowledgments: The authors thank to the following researchers and technicians who contributed to the sequencing effort: A. Dalben, A.L.A. Beraldo, A.R de Oliveira, A.S. Zanca, A.S. Castro, D. Truffi, E.A Amaral da Silva, E.A.N. Pedrinho, E.S. Ferro, E.L. da Silveira, F.S. Prada, G.H. Goldman, J.C. Setúbal, J.P. Piazza, K.M. Borges, K.M. Brito, L.B.D. Labuto, M.M. Zerillo, M.A.C. da Silva, M.C. Oliveira, C.R. Borges Neto, R.L.B.C. Oliveira, R. Padovani, Z.A.R. Souza. This project was sponsored by Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café (CBP&D-Café), Empresa Brasileira de Pesquisa Agropecuária (Embrapa) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
Ribas, A.F., Kobayashi, A.K., Pereira, L.F.P. and Vieira, L.G.E. (2006). Production of herbicide-resistant coffee plants (Coffea canephora P.) via Agrobacterium tumefaciens-mediated transformation. Brazil. Arch. Biol. Technol. (accepted for publication).
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombie WR, Venter JC (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252:1651-1656.
- Aldwinckle SH, Gaitan AL (2002) Constitutive and inducible promoters from coffee plants. [US Patent N° 6,441,273].
- Aldwinckle SH, Gaitan AL, (2005) Constitutive a-tubulin promoter from coffee plants and uses thereof. [US Patent N° 6903247].
- Altschul SF, Gish W, Miller W, Myers EW, Lipman, DJ (1990) Basic local alignment search tool. J. Mol. Biol. 215:403-10.
- Andrade AE, Albuquerque EVS, Grossi de Sá MF, Carneiro RMDG, Metha, A (2005) Expressão diferencial de proteínas em raízes de Coffea canephora infectadas com o nematóide endoparasita Meloidogyne paranaensis In: Anais do IV Simpósio de Pesquisa dos Cafés do Brasil. Londrina, Brasil. Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café, CD-ROM.
- Arruda P (2001) Sugarcane transcriptome: a landmark in plant genomics in the tropics. In: Arruda P (ed), Special volume on Sugarcane Transcriptome. Genet. Mol. Biol. 24:1296.
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9:208-218.
- Bertrand B, Anthony F, Lashermes P (2001) Breeding for resistance to Meloidogyne exigua in Coffea arabica by introgression of resistance genes of Coffea canephora. Plant Pathol. 50:637-643.
- Brandalise M, Maluf M.P, Guerreiro Filho O,Gonçalves W, Maia IG (2005) Caracterização de genes com expressão tecido específica em raízes e folhas de Coffea arabica In: Anais do IV Simpósio de Pesquisa dos Cafés do Brasil. Londrina, Brasil,Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café, CD-ROM.
- Bridson DM, Verdcourt B (1988) Flora of tropical East Africa: Rubiaceae. (Part 2). Cape Town: Iziko Museums of Cape Town, pp.415-747.
- Charrier A, Berthaud J (1985) Botanical classification of coffee. In: Clifford MN, Wilson KC (eds), Coffee: botany, biochemistry and production of beans and beverage, pp.13-47. Croom Helm, London, Sydney.
- Chenwei, L, Mueller, LA, Mc Carthy, J, Crouzillat, D, Pétiard, V, Tanksley, SD (2005). Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts, Theor. Appl. Genet. 112:114-130.
- Chevalier A, Dagron M (1928) Recherches historiques sur les débuts de la culture du caféier en Amérique. Communications et Actes de Ácadémie des Sciences Coloniales, Paris.
- Carvalho A, Fazuoli LC (1993) O melhoramento de plantas no Instituto Agronômico. In: Furlani AMC, Viégas GP (eds), Café. pp.29-76. Campinas, Brasil
- Carvalho A (1945) Distribuição geográfica e classificação botânica do gênero Coffea com referência especial à espécie Arabica. Bol. Superint. Serv. Café. 21:174-180.
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186194.
- Ewing B, Hillier L, Wend MC, Green P (1998) Basecalling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175185.
- Gaspari-Pezzopane C, Maluf MP, Pinto FO (2005) Expressão gênica diferencial em frutos de Coffea arabica L. em diferentes estádios de desenvolvimento e maturação. In: Anais do IV Simpósio de Pesquisa dos Cafés do Brasil. Londrina, Brasil, Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café, CD-ROM.
- Geromel C, Ferreira LP, Cavalari AA, Pereira LFP, Vieira LGE, Leroy T, Mazzafera P, Marraccini, P (2005) Metabolismo de açúcares durante o desenvolvimento de frutos de café. In: Anais do IV Simpósio de Pesquisa dos Cafés do Brasil. Londrina, Brasil, Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café, CD-ROM.
- Guerreiro Filho O, Medina Filho HP, Carvalho A (1991) Fontes de resistência ao bicho-mineiro. Perileucoptera coffeella em Coffea sp. Bragantia 50:45-55.
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A Systematic Proteomic Study of Seed Filling in Soybean. Establishment of High-Resolution Two-Dimensional Reference Maps, Expression Profiles, and an Interactive Proteome Database Plant Physiol. 137:1397-1419.
- Hatanaka T, Choi YE, Kusano T, Sano H (1999) Transgenic plants of Coffea canephora from embryogenic callus via Agrobacterium tumefaciens-mediated transformation. Plant Cell Rep. 19:106-110.
- Herrera JC, Combes MC, Cortina H, Lashermes P (2004) Factors influencing gene introgression into the allotetraploid Coffea arabica L. from its diploid relatives. Genome 47:1053-1060.
- Höfte H, Desprez T, Amselem J, Chiapello H, Caboche M, Moisan A, Jourjon MF, Charpenteau JL, Berthomieu P, Guerrier D, Giraudat J, Quigley F, Thomas F, Yu DY, Mache R, Raynal M, Cooke R, Grellet F, Delseny M, Parmentier Y, Marcillac G, Gigot C, Fleck J, Philipps G, Axelos M, Bardet C, Tremousaygue D, Lescure B (1993) An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana. Plant J. 4:1051-1061.
- Huang X, Madan A (1999) CAP3: A DNA assembly program. Genome Res. 9:868-877.
- Khun E (2001) From library screening to microarray technology: Strategies to determine gene expression profiles and to identify differentially regulated genes in plants. Ann. Bot. 87:139-155.
- Krug CA, Mendes JET, Carvalho A (1938) Taxonomia de Coffea arabica L. Descrição das variedades e formas encontradas no Estado de São Paulo. Boletim Técnico do Instituto Agronômico. Campinas, Brasil, 62:1-57.
- Lashermes P, Cros J, Marmey P, Charrier A (1993) Use of random amplified DNA markers to analyze genetic variability and relationships of Coffea species. Genet. Res. Crop Evol. 40:9199.
- Lashermes P, Cros J, Combes MC, Trouslot P, Anthony F, Hamon S, Charrier A (1996) Inheritance and restriction fragment length polymorphism of chloroplast DNA in the genus Coffea L. Theor. Appl. Genet. 93:626-632.
- Lashermes P, Combes MC, Trouslot P, Charrier A (1997) Phylogenetic relationships of coffee-tree species (Coffea L.) as inferred from ITS sequences of nuclear ribosomal DNA. Theor. Appl. Genet. 94:947-955.
- Leroy T, Henry AM, Royer M, Altosar I, Frutos R, Duris D, Philippe R (2000) Genetically modified coffee plants expressing the Bacillus thuringiensis cry1Ac gene for resistance to leaf miner. Plant Cell Rep. 19:382-389.
- Leroy T, Marraccini, P, Dufour, M, Montagnon, C, Lashermes, P., Sabau, X., Ferreira L.P., Jourdan, I., Pot, D., Andrade A. C., Glaszmann, J.C., Vieira, L.G. E. and Piffanelli P. (2005). Construction and characterization of a Coffea canephora BAC library to study the organization of sucrose biosynthesis genes. Theor. Appl. Genet. 111:10321041.
- Lockhart DJ, Winzeler EA (2000) Genomics, gene expression and DNA arrays. Nature, 405:827836.
- Maluf MP, Silvestrini M, Ruggiero LCM, Guerreiro Filho O, Colombo CA (2005) Genetic diversity of cultivated Coffea arabica inbred lines assessed by RAPD, AFLP and SSR marker systems. Sci. Agric. 62:366-373.
- Marraccini P, Deshayes A, Petiard V, Rogers WJ (1999) Molecular cloning of the complete 11S seed storage protein gene of Coffea arabica and promoter analysis in transgenic tobacco plants. Plant Physiol. Biochem. 37:273-282.
- Marraccini P, Courjault C, Caillet V, Lausanne F, Lepage B, Rogers WJ, Tessereau S, Deshayes A (2003) Rubisco small subunit of Coffea arabica: cDNA sequence, gene cloning and promoter analysis in transgenic tobacco plants. Plant Physiol. Biochem. 41:17-25.
- Noir S, Patheyron S, Combes MC, Lashermes P, Chalhoub B (2004) Construction and characterization of a BAC library for genome analysis of the allotetraploid coffee species (Coffea arabica L.). Theor. Appl. Genet. 109:225-230.
- Ogita S, Uejuji H, Yamaguchi Y, Koizumi N, Sano H (2003) RNA interference: Producing decaffeinated coffee plants. Nature 423:823.
- Pereira A (2000) A transgenic perspective on plant functional genomics. Transgenic Res. 9:245260.
- Pearl HM, Nagai C, Moore PH, Steiger DL, Osgood RV, Ming R (2004) Construction of a genetic map for arabica coffee. Theor. Appl. Genet. 108:829835.
- Prakash NS, Combes MC, Somanna N, Lashermes P (2002) AFLP analysis of introgression in coffee cultivars (Coffea arabica L.) derived from a natural interspecific hybrid. Euphytica 124:265-271.
- Prakash NS, Marques DV, Varzea VMP, Silva MC, Combes MC, Lashermes P (2004) Introgression molecular analysis of a leaf rust resistance gene from Coffea liberica into C. arabica L. Theor. Appl. Genet. 109:1311-1317.
- Ribas AF, Kobayashi AK, Pereira LFP, Vieira, LGE (2005) Genetic transformation of Coffea canephora P. by particle bombardment. Biol. Plant. 49:493-497.
- Rounsley SD, Glodek A, Sutton G, Adams MD, Somerville CR, Venter JG, Kerlavage AR (1996) The construction of Arabidopsis expressed sequence tag assemblies. Plant Physiol. 112:1177-1183.
- Rufino RJN, Caixeta ET, Zambolim EM, Pena GF, Almeida RF, Alavarenga SM, Zambolim L, Sakaiyama NS (2005) Microsatellite markers for coffee tree. In: Anais do IV Simpósio de Pesquisa dos Cafés do Brasil. Londrina, Brasil.Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café. CD-ROM.
- Sales RMOB, Andrade AC, da Silva FR (2005) Determinação do Unigene do Projeto Genoma Café. In: Anais do IV Simpósio de Pesquisa dos Cafés do Brasil. Londrina, Brasil Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café. CD-ROM.
- Satyanarayana KV, Kumar V, Chandrashekar A, Ravishankar GA (2005) Isolation of promoter for N-methyltransferase gene associated with caffeine biosynthesis in Coffea canephora J. Biotech. 119:2025.
- Sera T, Ruas PM, Ruas CD, Diniz LEC, Carvalho VD, Rampim L, Ruas EA, Silveira SR (2003) Genetic polymorphism among 14 elite Coffea arabica L. cultivars using RAPD markers associated with restriction digestion. Genet. Mol. Biol. 1:59-64.
- Telles GP, da Silva FR (2001) Trimming and clustering sugarcane ESTs. Gen. Mol. Biol. 24:17-23.
- Vincent D, Lapierre C, Pollet B, Cornic G, Negroni L, Zivy M (2005) Water deficits affect caffeate O-methyltransferase, lignification, and related enzymes in maize leaves. A proteomic investigation. Plant Physiol. 137: 949-960.
- Vinecky F, Brito KM, da Silva FR, Andrade AC (2005) Análise in silico de genes potencialmente envolvidos na resposta aos estresses abióticos presentes na base de dados do Genoma Café. In: Anais do IV Simpósio de Pesquisa dos Cafés do Brasil. Londrina, Brasil. Consórcio Brasileiro de Pesquisa e Desenvolvimento do Café. CD-ROM.
- Yamamoto K, Sasaki TL (1997). Large-scale EST sequencing in rice. Plant Mol. Biol. 35:135-144.
- Wolfsberg TG, Landsman D (1997) A comparison of expressed sequence tags (ESTs) to human genomic sequences. Nucleic Acids Res. 25:1626-1632.
Publication Dates
-
Publication in this collection
20 June 2006 -
Date of issue
Mar 2006