Abstract
Due to the development of antibiotic resistance in microorganisms, antimicrobial peptides from natural sources have attracted attention in recent times. Several antimicrobial peptides have been isolated from a wide range of animal sources, particularly snake venoms. Naja naja venom showed antibacterial as well as direct and indirect hemolytic activities, and an antibacterial peptide was purified through gel permeation and ion exchange chromatography. Its molecular mass was 2491Da, which was determined using Matrix Assisted Laser Desorption/Ionization-Time-of-Flight (MALDI-TOF) mass spectrometry and the amino acids sequence of the N-terminus was DEQSTHGAYVWKL. The purified peptide showed potent antibacterial activity against Gram-negative and Gram-positive bacterial strains like Escherichia coli, Pseudomonas aeruginosa and Vibrio cholerae, and Staphylococcus aureus, Enterococcus faecalis, Streptococcus pneumoniae, Streptococcus pyogenes, Bacillus subtilis, respectively. The most potent activity was towards Gram-negative bacteria. Activity was retained at concentrations as low as 100µg/ml. Minimum inhibitory concentrations (MIC; in mg) of Naja Antibacterial Peptide (NAP) and known antibiotics against Gram-positive and Gram-negative bacteria were determined using microdilution susceptibility test in sterile 96-well microdilution plates. However, the peptide did not show direct or indirect hemolytic activity.
Indian cobra; Naja naja; snake venom antibacterial peptide; hemolytic activity
ORIGINAL PAPER
Characterization of an antibacterial peptide from indian cobra (Naja naja) venom
Sachidananda M. K.I; Murari S. K.II; Channe Gowda D.III
IDepartment of Chemistry, University of Mysore, Manasagangotri, Mysore, India
IIDepartment of Biochemistry, University of Mysore, Manasagangotri, Mysore, India
Correspondence to Correspondence to: Doddhalli Channe Gowda Department of Studies in Chemistry University of Mysore Manasagangothri Mysore 570 006, Karnataka, India Phones: +91 821 2515525 Ext. 40(Office) and +91 821 2340011(Residence) Fax: +91 821 2421263 Email: getsachi@rediffmail.com
ABSTRACT
Due to the development of antibiotic resistance in microorganisms, antimicrobial peptides from natural sources have attracted attention in recent times. Several antimicrobial peptides have been isolated from a wide range of animal sources, particularly snake venoms. Naja naja venom showed antibacterial as well as direct and indirect hemolytic activities, and an antibacterial peptide was purified through gel permeation and ion exchange chromatography. Its molecular mass was 2491Da, which was determined using Matrix Assisted Laser Desorption/Ionization-Time-of-Flight (MALDI-TOF) mass spectrometry and the amino acids sequence of the N-terminus was DEQSTHGAYVWKL. The purified peptide showed potent antibacterial activity against Gram-negative and Gram-positive bacterial strains like Escherichia coli, Pseudomonas aeruginosa and Vibrio cholerae, and Staphylococcus aureus, Enterococcus faecalis, Streptococcus pneumoniae, Streptococcus pyogenes, Bacillus subtilis, respectively. The most potent activity was towards Gram-negative bacteria. Activity was retained at concentrations as low as 100µg/ml. Minimum inhibitory concentrations (MIC; in mg) of Naja Antibacterial Peptide (NAP) and known antibiotics against Gram-positive and Gram-negative bacteria were determined using microdilution susceptibility test in sterile 96-well microdilution plates. However, the peptide did not show direct or indirect hemolytic activity.
Key words: Indian cobra, Naja naja, snake venom antibacterial peptide, hemolytic activity.
INTRODUCTION
Antimicrobial Peptides (AMPs) are an extremely diverse group of small proteins that have in common a native antimicrobial activity. The existence of AMPs has been known for several decades, but only recently has their function been recognized as essential to the animal immune response. They participate primarily in the innate immune system and are used as a first line of immune defense by many organisms, including plants, insects, bacteria and vertebrates (9). These molecules are peptides with a high level of basic and hydrophobic amino acids. They present a broad antimicrobial spectrum against bacteria, fungi or parasites, by acting through insertion into the cell membrane or bind to receptors. Therefore, these molecules are promising for the development of antibiotics especially for the treatment against multiresistant microorganisms (7, 19). For this reason, significant commercial interest and effort have been made to develop cationic peptides as potential antimicrobial therapeutics. Recent studies have clarified that antimicrobial peptides are an important component of the innate defense of all species (5).
More than 700 AMPs have already been identified in living species like bacteria, fungi, amphibians, insects, reptiles and mammals (10). In the last years, several AMPs have been found in different venoms from different animals and are traditionally linked to defense mechanisms (6). Snake venoms are rich sources of pharmacologically active polypeptides and proteins. Peptides from snake venoms are of biological interest as a potential source of active compounds. These molecules could act as (or be used as a prototype for) (i) therapeutic agents; (ii) research tools for the diagnosis of several diseases; (iii) basic research about physiological and pathological processes (22).
Snake venom has been established to show bactericidal activity (8) and the action of its proteins on E. coli has been extensively studied (23). The action of snake venom AMPs on clinical bacterial strains has also been reported (24). Venoms from 30 different snake species were tested using disk diffusion test for antibacterial activity (21). In the present study, the isolation and characterization of a peptide from N. naja venom was reported as well as its inhibitory activity against different bacterial strains.
MATERIALS AND METHODS
Materials
Indian cobra (Naja naja) venom was purchased from Irulla Snake-Catchers Association, Chennai, Tamil Nadu, India. CM-Sephadex C-25, Sephadex G-25, Sephadex G-75, and Bovine Serum Albumin (BSA) were purchased from Sigma Chemical Company, St. Louis, MO, USA. The bacterial strains E. coli American Type Culture Collection (ATCC) 25922, E. coli ATCC 476, S. aureus National Cell Type Cell Culture (NCTCC) 6570, S. aureus NCTCC 6571, P. aeruginosa ATCC 26519, P. aeruginosa NCTCC 10662, V. cholerae Wild strain, E. faecalis MTCC 459, E. faecalis MTCC 439, S. pneumoniae MTCC 497, S. pneumoniae MTCC 7978, S. pyogenes NCTCC 7465, S. pyogenes NCTCC 7978, B. subtilis NCTCC 1040, and B. subtilis NCTCC 8236 were purchased from American Type Cell Culture Institute, USA, and Institute of Microbial Technology, Chandighar, India. Human blood samples were collected from healthy volunteers from the Department of Biochemistry, University of Mysore, Mysore, India. All other chemicals used were of analytical grade. All the solvents were redistilled before use.
Protein Estimation
Protein concentration was determined according to the method of Lowery et al. (16) using BSA as standard.
Sephadex G-75 Column Chromatography
Lyophilized N. naja venom (300mg in 1ml) was dissolved in 10mM potassium phosphate buffer, pH 7.4, and centrifuged at 5000g for 5min. The supernatant was applied to a column (0.8cm X 120cm) of Sephadex G-75 equilibrated and eluted with the same buffer at 20ºC. The fractions from the column were eluted at a flow rate of 20ml/h and 2ml fractions were collected. Protein elution was monitored at 280nm using a Shimadzu spectrophotometer (1601A). Alternate tubes were assayed for antibacterial activity. Fractions presenting activity were individually pooled, desalted, lyophilized and stored at -4ºC.
CM-Sephadex C-25 Column Chromatography
Peak III (90mg in 3ml equilibrating buffer) from Sephadex G-75 column was loaded onto a CM-Sephadex C-25 column (1.2cm X 40cm) equilibrated with 10mM potassium phosphate buffer, pH 7.4. The column was eluted by a stepwise gradient of potassium phosphate buffer (10mM150mM) and NaCl (0.01M1M). Fractions were eluted at 20ºC at a flow rate of 25ml/h and 2.5ml fractions were collected. Protein elution was monitored at 280nm using a Shimadzu spectrophotometer. Fractions presenting antibacterial activity were pooled, desalted, lyophilized and stored at -4ºC.
Sephadex G-25 Column Chromatography
Peak IV (36mg in 1ml of equilibrating buffer) from CM-Sephadex C-25 column was loaded onto a Sephadex G-25 column (0.75cm X 60cm) equilibrated with 10mM potassium phosphate buffer, pH 7.4. The fractions from the column were eluted at a flow rate of 20ml/h and 2ml fractions were collected. Protein elution was monitored at 280nm using a Shimadzu spectrophotometer (1601A). Alternate tubes were assayed for antibacterial activity. Fractions presenting activity were individually pooled, desalted, lyophilized and stored at -4ºC.
High-Performance Liquid Chromatography
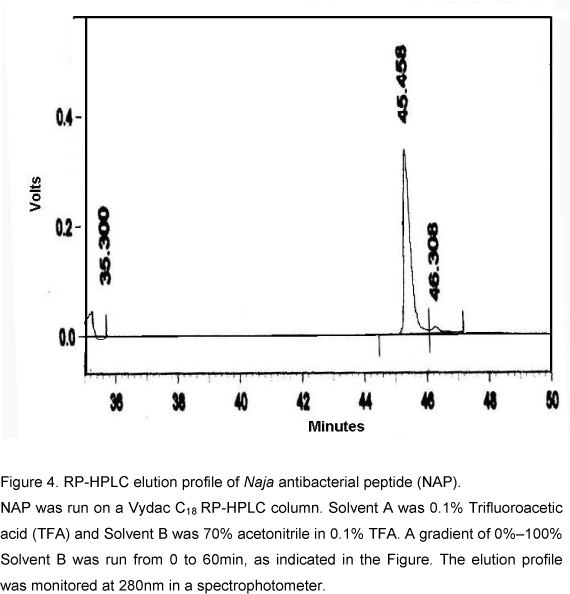
Purified NAP was subjected to Reverse Phase-High-Performance Liquid Chromatography (RP-HPLC) on Vydac-C18 (5µm, 0.21cm X 25cm) column. The column was first equilibrated with Solvent A (0.1% Trifluoroacetic Acid TFA) until the base line monitored at 220nm was stable. The peptide was then injected into the column. Elution was carried out with a linear gradient (0%100%) of Solvent B (70% Acetonitrile in 0.1% TFA). Peptide elution was monitored at 220nm.
Mass Spectrometry
The molecular mass of NAP was determined using MALDI-TOF mass spectrometry (Voyager Spec # 1 MC) in positive ionization mode. a-Cyano-4-hydroxycinnamic acid was used as MALDI matrix.
N-Terminal Sequencing
The terminal sequencing of NAP was carried out in a fully automated Shimadzu protein sequencer (PSQ-1) system that employs Edman's degradation reaction for sequential separation of N-terminal amino acids.
Hemolytic Activity
Direct and indirect hemolytic activities were assayed as described by Boman and Kalletta (4). The substrate for direct hemolytic activity was prepared by suspending 1ml of packed fresh human red blood cells (RBC) in 9ml of phosphate buffered saline (PBS). For indirect hemolytic activity, 1ml of fresh hen's egg yolk was included in the above-mentioned suspension. One ml of the suspension was incubated with different concentrations of NAP for 45min at 37°C. The reaction was stopped by adding 9ml of ice-cold PBS. The suspensions were centrifuged at 2000g for 20min and the released hemoglobin was read at 530nm. The activity was expressed as percentage (%) of hemolysis.
Evaluation of Antibacterial Activity
Antibacterial assay was described by Linzxing Zhong et al. (15). The microorganisms were grown in Muller-Hinton broth. After incubation for 1618h at 37ºC, the bacteria were harvested by centrifugation (2000g for 10min), washed twice with 10mM sodium phosphate buffer, pH 6.0, and finally resuspended in 10ml buffer. Its density was determined by measuring the absorbance at A600. The peptide MIC was determined using a microdilution susceptibility test in sterile 96-well microdilution plates. Microorganisms (1X104 to 2X104 CFU/50l) were pipetted into the wells, which consists of 20-300g of peptide/ml. Assays for peptide were performed in duplicate with each bacterium. After 24h incubation at the optimal growing temperature, the optical density (OD) at 600nm was read on an absorption microtiter plate reader (Biotek Instruments INC.). The percentage of inhibition was calculated as {1-(a\b) X 100}, where a = OD 600nm of the bacteria with peptide, and b = OD 600nm of the control well containing only buffer, bacteria and media. The MIC evaluation was defined as 100% inhibition. Control was run by replacing the peptide solution with buffer solution. Respective antibiotics were used as standard drugs replacing peptide solution.
Statistics
For all experiments, results were expressed as the mean ± SEM of at least 3 independent experiments.
RESULTS
Purification of NAP
Naja naja venom (300mg) subjected to gel permeation chromatography on a Sephadex G-75 column resolved into three protein peaks. The antibacterial activity of the whole venom and its fraction is shown in Table 1. When all the peaks were screened for antibacterial activity, only peak III showed activity (Fig. 1). The peak III from the Sephadex G-75 fraction was pooled, concentrated and desalted using Sephadex G-10 column. The pooled peak III fraction was further resolved into four peaks on CM-Sephadex C-25 column by applying NaCl gradient (Fig. 2). Only peak IV exhibited potent antibacterial activity and contributed to 73.98% of the total activity loaded, and 4.5% of the protein loaded on CM-Sephadex C-25 column was recovered in this antibacterial fraction. The antibacterial activity of this fraction was increased by 6-fold compared to the whole venom activity. Further, peak IV from the CM-Sephadex C-25 fraction was pooled, concentrated and loaded onto Sephadex G-25 column. On fractionation, the peptide components resolved into two peaks, which were designated as peak A and peak B (Fig. 3). Peak B showed significant antibacterial activity and increased by 10-fold, compared to the whole venom activity.
The homogeneity of the antibacterial peptide was examined by RP-HPLC using C18 column. The elution buffer contained 0.1% TFA and was eluted with acetonitrile gradient. NAP eluted as single symmetrical sharp peak with a retention time of 43.6min (Fig. 4). The molecular mass determined by MALDI-TOF mass spectrometry was 2491Da (Fig. 5). The NAP N-terminal sequence analysis indicated the following 13 amino acids sequence: DEQSTHGAYVWKL. All these data clearly established that the antibacterial peptide was purified to homogeneity.
Antibacterial Activity of NAP
The isolated peptide was tested against four species of Gram-positive and three species of Gram-negative bacterial strains. NAP was also tested against a wide variety of Gram-positive and Gram-negative bacteria collected from immunosuppressed patients following disease or chemotherapy or from patients suffering from other chronic diseases.
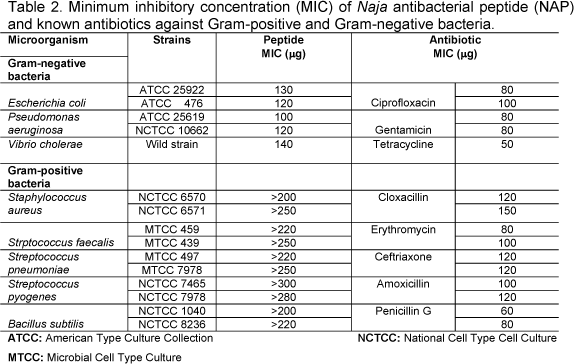
NAP exhibited antibacterial activity against a variety of bacterial clinical isolates; this was evaluated by determining the MIC values. The difference in the activity of NAP between Gram-positive and Gram-negative is well marked. It was more effective among Gram-negative bacteria in general, and not all species were susceptible (Table 2). Staphylococcus aureus, Staphylococcus faecalies, Streptococcus pneumoniae, and Streptococcus pyogenes showed MIC values > 300µg/ml. On the other hand, E. coli, Pseudomonas aeruginosa, Vibrio cholerae, and Klebsilla pneumoniae were more susceptible to NAP. Hemolytic activity of whole venom exhibited direct and indirect lytic activity on human RBC causing 83% hemolysis. Similarly, NAP was incubated with washed RBC at 37°C for 10min and increased in dose-dependent manner. NAP did not show direct or indirect hemolytic activity even at higher concentrations.
Sephadex G-75 column (0.8cm X 120cm) was eluted with 10mM potassium phosphate buffer, pH 7.4, at a flow rate of 20ml/h and 2ml fractions were collected. The protein elution profile () was monitored at 280nm in a spectrophotometer. Fractions showing antibacterial activity (dotted line) were pooled for further fractionation.
The column (1.2cm X 40cm) was pre-equilibrated with 10mM potassium phosphate buffer, pH 7.4, and eluted by a stepwise gradient of potassium phosphate buffer (10mM150mM) and NaCl (0.01M1M) as indicated on the top of the Figure; 2.5ml fractions were collected and the proteins elution profile () was monitored at 280nm in a spectrophotometer. Fractions showing antibacterial activity (dotted line) were pooled and desalted for further homogeneity.
Sephadex G-25 column (0.75cm X 60cm) was eluted with 10mM potassium phosphate buffer, pH 7.4, at a flow rate of 20ml/h and 2ml fractions were collected. The protein elution profile () was monitored at 280nm in a spectrophotometer. Fractions showing antibacterial activity (dotted line) were pooled for further analysis.
NAP was run on a Vydac C18 RP-HPLC column. Solvent A was 0.1% Trifluoroacetic acid (TFA) and Solvent B was 70% acetonitrile in 0.1% TFA. A gradient of 0%100% Solvent B was run from 0 to 60min, as indicated in the Figure. The elution profile was monitored at 280nm in a spectrophotometer.
DISCUSSION
Several AMPs have been found in different venoms from different animals and are traditionally linked to defense mechanisms (6). Antimicrobial peptides have an ability to kill or neutralize Gram-negative and Gram-positive bacteria, fungi (including yeasts), parasites (including planaria and nematodes), cancer cells, and enveloped viruses like HIV and herpes simplex virus (11). In this study, the low-molecular-weight peptide from snake venom was referred as NAP. For the first time, purification and N-terminal sequencing of a new potent antibacterial peptide from Naja naja snake venom was reported. The peptide was isolated from N. naja whole venom by subjecting it to gel permeation and ion exchange chromatography, which resulted in 10-fold purification. RP-HPLC, MALDI-TOF and N-terminal sequencing analysis confirmed NAP homogeneity. Based on Basic Local Alignment Search Tool (BLAST) of the primary N-terminal sequence of the antibacterial peptide, NAP is different from the antibacterial peptides reported so far.
It is generally accepted that different venoms have several thousands of proteins with different properties. However, in recent years, more than 700 cationic peptides have been isolated from mammals, amphibians, reptiles, arthropods, plants, bacteria and viruses (3,7,19). Some of the first reports about antibacterial activity in snake venoms were in 1948 and in 1968, involving Elapidae and Viperidae venoms (8, 25). Venoms from snakes of the Viperidae family present antimicrobial activity against Sarcina species, while in the Elapidae family, a lytic factor or cytotoxin composed of a basic low-molecular-weight protein was found in Naja species, and Hemachatus haemachatus was shown to have antibacterial activity. They were able to disrupt S. aureus and E. coli phospholipid membranes, respectively (23, 25). In the present study, NAP displayed higher inhibitory activity against Gram-negative bacteria like E. coli, P. aeruginosa, V. cholerae, than against Gram-positive bacteria like S. aureus, S. faecalis, S. pneumoniae, S. pyogenes and B. subtilis.
The peptide dissolves divalent cations that are essential for outer membrane and consequently distorts the outer membrane bilayer (20). This allows access to the cytoplasmic membrane where peptides channel formation has been proposed to occur (13). It is an intermediate step in the uptake of peptides into the cytoplasm, where it inhibits an essential function by binding to polyanionic DNA (18,26). It has been argued that antimicrobial peptides provides organisms with molecules that are rapidly synthesized because of small size, less costly to synthesize than antibodies or specific phagocytic cells, and can be stored if necessary as processed biologically-active components which are rapidly available for host defense (2). The ability of the antibacterial peptide to lyse cells is the result of a complex interrelationship of factors involving conformation, charge, hydrophobic and amphipathicity. The cationic residues in an antimicrobial peptide are considered to be important in the initial binding to the negatively charged phospholipids in the cell membranes of microorganisms (27). It has been suggested that increasing the hydrophobic moment of an antimicrobial peptide has a relatively modest effect on the ability to make the microorganisms negatively-charged cell membrane permeable but not a marked effect on the more zwitterionic phospholipid membrane of the erythrocyte (26). In general, although it is accepted that a polypeptide chain of at least 20 amino acids is necessary to span the membrane lipid bilayer to effect the formation of ion channels (14), shorter cationic alpha-helical amphipathic peptides of 812 residues can also form ion channels, presumably through head-to-tail dimerization of the peptides (1).
In a conventional assay on human RBC, whole venom caused significant hemolysis, but NAP did not cause a significant hemolysis. It was known that appearance of numerous contiguous apolar residues in a helix is necessary for a significant hemolysis to occur (17). Like other antimicrobial peptides, the polar residues in the NAP might be well interspersed among the hydrophobic residues, interrupting the contiguity of hydrophobicity, which gives the potential to form an amphipathic helix. For this reason, NAP probably exhibits little hemolytic activity like many other antimicrobial activities. The widespread use of antibiotics has caused numerous antibiotic resistant strains to develop, resulting in the continuous need for new antibiotics. Studies directed towards understanding the relationships between the secondary structure and biological activities of these natural peptides have indicated that amphipatic alpha-helical conformation plays an important role in their biological activities (12). In conclusion, the present study on N. naja venom suggests the presence of a potent antibacterial peptide. Further studies on this peptide would be interesting, the clinical isolates were investigated can cause infections at sites where treatment with this type of peptide would probably help in the development as a potential therapeutic agent applicable for clinical isolates.
ACKNOWLEDGMENTS
We acknowledge Department of Biochemistry and Biotechnology, University of Mysore, Manasagangothri, Mysore, India, for providing instrumentation facility.
Received: April 25, 2006
Accepted: March 12, 2007
Abstract published online: March 19, 2007
Full paper published online: May 31, 2007
Conflicts of interest: There is no conflict
Financial source: University grants commission, New Delhi.
- 1 AGAWA Y., LEE S., ONO S., AOYOGI H., OHNO M., TANNIGUCHI T., ANZAI K., KIRINO Y. Interaction with phospholipid bilayers, ion channel formation, and antimicrobial activity of basic amphipathic alpha-helical model peptides of various chain lengths. J. Biol. Chem, 1991, 266, 20218-22.
- 2 BOMAM HG. Antibacterial peptides: key components needed in immunity. Cell, 1991, 65, 205-7.
- 3 BOMAN HG. Antibacterial peptides: basic facts and emerging concepts. J. Int. Med., 2003, 254, 197-215.
- 4 BOMAN HG., KALLETTA U. Chromatography of rattlesnake venom: a separation of three phosphodiesterases. Biochem. Biophys. Acta, 1957, 24, 619-24.
- 5 GABAY JE. Ubiquitous natural antibiotics. Science, 1994, 264, 373-4.
- 6 GALLO RL., MURKAMI M., OHTAJE T., ZAIOU M. Biology and clinical relevance of naturally occurring antimicrobial peptides. J. Allergy Clin. Immunol, 2002,110, 823-31.
- 7 GANZ T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol., 2003, 3, 710-20.
- 8 GLASER HRS. Bactericidal activity of Crotalus venom in vitro Copeia, 1948, 4, 245-7.
- 9 GOMES VM., CARVALHO AO., CUNHA M., KELLER MN., BLOCH JR C., DEOLINDO P., ALVES EW. Purification and characterization of a novel peptide with antifungal activity from Bothrops jararaca venom. Toxicon, 2005, 45, 817-27.
- 10 HANCOCK RE., CHAPPLE D. Peptide antibiotics. Antimicrob. Agents Chemother, 1999, 43, 1317-23.
- 11 HANCOCK RE., SCOOT MG. The role of antimicrobial peptides in animal defences. Proc. Natl. Acad. Sci. USA, 2000, 97, 8856 -62.
- 12 KAISER ET., KEZDY FJ. Peptides with affinity for membranes. Ann. Rev. Biophys. Biophys. Chem, 1987, 16, 561-81.
- 13 LEAR JD., WASSERMAN ZR., GRADO WF. Synthetic amphiphilic peptide models for protein ion channels. Science, 1988, 240, 1177-81.
- 14 LEHRER RI., GANZ T., SALTED ME. Defesins: endogenous antibiotic peptides of animal cells. Cell, 1991, 64, 229-30.
- 15 LINZXIUG ZHONG., REBECCA PUTNAM J., CURTIS JOHNSON JR., GURAJ RAO A. Design and synthesis of amphipathic antimicrobial peptides. Int. J. Pept. Protein Res, 1995, 45, 337-47.
- 16 LOWRY OH., ROSEBROUGH NJ., FARR AL., RANOALL RJ. Protein measurement with the Folin-Phenol reagent. J. Biol. Chem, 1951, 193, 265-75.
- 17 NICOLAS P., MOR A. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Ann. Rev. Microbiol, 1995, 49, 277-304.
- 18 PARK CB., KIM HS., KIM SC. Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun, 1998, 224, 253-7.
- 19 PEREZ-TRALLERO E., IGLESIAS L. Tetracyclines, sulfonamides and metronidazole. Enferm. Infecc. Microbiol. Clin., 2003, 21, 520-9.
- 20 PETERSON AA., FESIK SW., MCGROARTY EJ. Decreased binding of antibiotics to lipopolysaccharides from polymyxin-resistant strains of Escherichia coli and Salmonella typhimurium Antimicrob. Agents Chemother, 1987, 31, 230-7.
- 21 STILES BG., SEXTON FW., WEINSTEIN SA. Antibacterial effects of different snake venoms: purification and characterization of antibacterial proteins from Pseudechis australis (Australian king brown or mulga snake) venom. Toxicon, 1991, 29, 1129-41.
- 22 STOCKER KF. Medical Use of Snake Venom Proteins Boca Raton: CRC Press, 1990.
- 23 STOCKER JF., TRAYNOR JR. The action of various venoms on Escherichia coli J. Appl. Bacteriol, 1986, 61, 383-8.
- 24 TALAN DA., CITRON DM., OVERTURN GD., SINGER B., FROMAN P., GOLDSTEIN EJ. Antibacterial activity of crotalid venoms against oral snake flora and other clinical bacteria. J. Infect. Dis, 1991, 164, 195-8.
- 25 WHITE J. Bites and stings from venomous animals: a global overview. Ther. Drug. Monit, 2000, 22, 65-8.
- 26 WIEPRECHT T., DATHER M., KRAUSE E., BEYERMANN M., MALOY WL., MACDONALD DL., BIENERT M. Modulation of membrane activity of amphipathic, antibacterial peptides by slight modifications of the hydrophobic moment. FEBS Lett, 1997, 417, 135-40.
- 27 YEAMAN MR., YOUNT NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev, 2003, 55, 27-55.
Publication Dates
-
Publication in this collection
13 June 2007 -
Date of issue
2007
History
-
Received
25 Apr 2006 -
Accepted
12 Mar 2007